Difference between revisions of "Electrodes and Surface Locations"
| Line 134: | Line 134: | ||
Auxiliary files can complement the information in the data file. Here we specify what happens when a data file is opened: | Auxiliary files can complement the information in the data file. Here we specify what happens when a data file is opened: | ||
| − | |||
'''1.''' If the data file has been read previously, the database entry specifies which auxiliary files should be read. The file and the specified auxiliary files are opened and the data are displayed. | '''1.''' If the data file has been read previously, the database entry specifies which auxiliary files should be read. The file and the specified auxiliary files are opened and the data are displayed. | ||
| Line 162: | Line 161: | ||
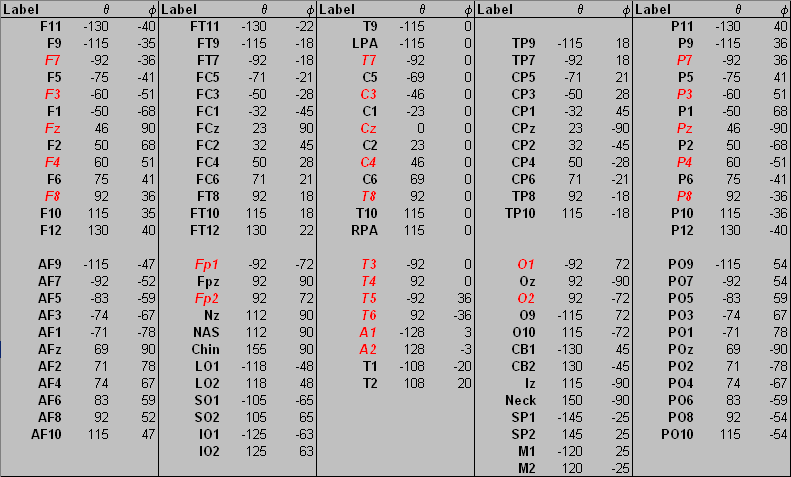
''Electrode labels in the file <span style="color:#ff9c00;">'''default.ecd '''</span>and their spherical coordinates. 10-20 electrodes are shown in<span style="color:#ff0000;"> red and italic.''</span> | ''Electrode labels in the file <span style="color:#ff9c00;">'''default.ecd '''</span>and their spherical coordinates. 10-20 electrodes are shown in<span style="color:#ff0000;"> red and italic.''</span> | ||
| − | |||
The spherical coordinates are defined in degrees by the azimuth (from Cz, positive = right, negative = left hemisphere) and the latitude (counterclockwise from T7/T3 for left and from T8/T4 for right hemisphere) of each electrode. Please do not modify the existing labels or coordinates in this file, because this would adversely affect the interpolated (virtual) montages, the maps and the source montages and source images in BESA Research. However, you may add additional labels for scalp electrodes at the end of this file if needed (up to a total of 196). When you edit the electrode configuration or read in electrode files, BESA Research may replace the 10/20 standard labels T3, T4, T5, T6 by their new 10/10 equivalents T7, | The spherical coordinates are defined in degrees by the azimuth (from Cz, positive = right, negative = left hemisphere) and the latitude (counterclockwise from T7/T3 for left and from T8/T4 for right hemisphere) of each electrode. Please do not modify the existing labels or coordinates in this file, because this would adversely affect the interpolated (virtual) montages, the maps and the source montages and source images in BESA Research. However, you may add additional labels for scalp electrodes at the end of this file if needed (up to a total of 196). When you edit the electrode configuration or read in electrode files, BESA Research may replace the 10/20 standard labels T3, T4, T5, T6 by their new 10/10 equivalents T7, | ||
| + | T8, P7, P8. However, in the initialization file <span style="color:#ff9c00;">'''BESA.ini '''</span>you can reset to the old 10/20 standard by relabeling T7=T3, P7=T5, T8=T4, P8=T6 under the heading [Electrodes]. You may use the same feature to assign appropriate labels to the X1..X8 channels which exist in many systems, e.g. X1=EKG1 etc. | ||
| − | |||
=== Recommendations for electrode placement === | === Recommendations for electrode placement === | ||
For source montages and source analysis two principles are important: | For source montages and source analysis two principles are important: | ||
| − | |||
# Covering of the lower head with inferior electrodes to record activity from the inferior surfaces of the brain, especially from the basal temporal lobe, from the temporal pole, from orbito-frontal cortex, and from basal occipital and cerebellar areas. | # Covering of the lower head with inferior electrodes to record activity from the inferior surfaces of the brain, especially from the basal temporal lobe, from the temporal pole, from orbito-frontal cortex, and from basal occipital and cerebellar areas. | ||
# Equal spacing of the electrodes over the whole head to cover all brain areas. | # Equal spacing of the electrodes over the whole head to cover all brain areas. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
In the following montage EEGxx the number xx indicates the number of electrodes. | In the following montage EEGxx the number xx indicates the number of electrodes. | ||
| − | |||
| − | |||
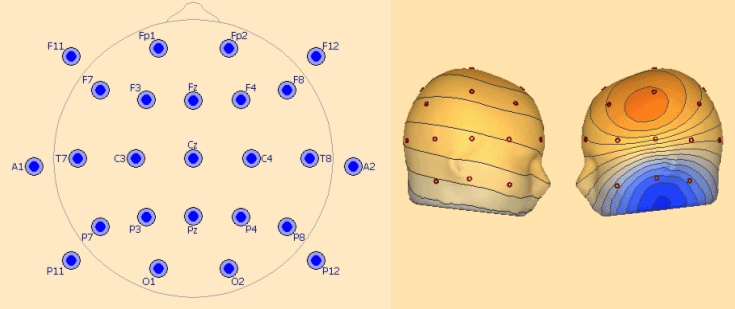
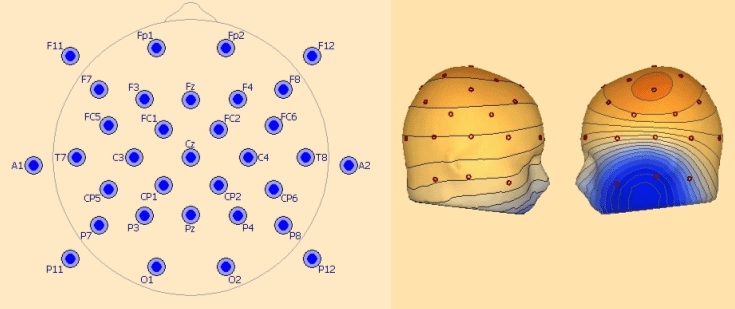
'''EEG25 - Minimum 10-20 configuration including inferior electrodes''' | '''EEG25 - Minimum 10-20 configuration including inferior electrodes''' | ||
| − | |||
This covers the 19 standard 10-20 electrodes: | This covers the 19 standard 10-20 electrodes: | ||
| Line 206: | Line 194: | ||
''Left: recommended configuration for 25 electrodes. Right: left temporal basal activity mapped with 25 electrodes.'' | ''Left: recommended configuration for 25 electrodes. Right: left temporal basal activity mapped with 25 electrodes.'' | ||
| − | |||
| − | |||
| Line 221: | Line 207: | ||
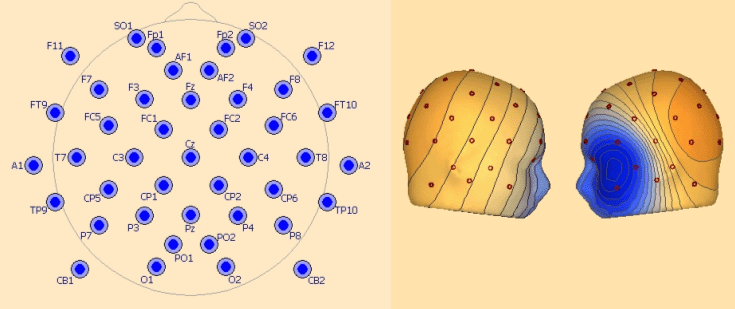
''Left: recommended configuration for 33 electrodes. Right: left temporal basal activity mapped with 33 electrodes.'' | ''Left: recommended configuration for 33 electrodes. Right: left temporal basal activity mapped with 33 electrodes.'' | ||
| − | |||
| − | |||
| Line 228: | Line 212: | ||
SO1, SO2 | SO1, SO2 | ||
| − | |||
| − | |||
| Line 237: | Line 219: | ||
F11, FT9, TP9, P11, F12, FT10, TP10, P12 | F11, FT9, TP9, P11, F12, FT10, TP10, P12 | ||
| − | |||
| − | |||
| Line 246: | Line 226: | ||
AF1, AF2, PO1, PO2 | AF1, AF2, PO1, PO2 | ||
| − | |||
| − | |||
| Line 266: | Line 244: | ||
With 64 or more channel caps, it is similarly recommended to use a sufficient number of inferior electrodes all around the head. At least 4 inferior temporal electrodes on each side and additional electrodes above or below the eyes (outside of the cap) are suggested. | With 64 or more channel caps, it is similarly recommended to use a sufficient number of inferior electrodes all around the head. At least 4 inferior temporal electrodes on each side and additional electrodes above or below the eyes (outside of the cap) are suggested. | ||
| + | |||
=== Editing the channel configuration === | === Editing the channel configuration === | ||
| Line 271: | Line 250: | ||
Only use the channel configuration editing facility if the electrodes or the common reference have not been correctly defined by your digital EEG system, or if you want to define specific spherical coordinates for your scalp electrodes. It is your responsibility to check and provide the correct sequence of electrode labels in correspondence with the sequence of channels in the EEG data file. If these sequences do not match exactly, errors will occur in the computation of maps, source images and interpolated montages. | Only use the channel configuration editing facility if the electrodes or the common reference have not been correctly defined by your digital EEG system, or if you want to define specific spherical coordinates for your scalp electrodes. It is your responsibility to check and provide the correct sequence of electrode labels in correspondence with the sequence of channels in the EEG data file. If these sequences do not match exactly, errors will occur in the computation of maps, source images and interpolated montages. | ||
| − | We will use the example EEG file <span style="color:#ff9c00;">'''eeg2.eeg'''</span> in the subdirectory ''Examples/EEG-Focus'' | + | We will use the example EEG file <span style="color:#ff9c00;">'''eeg2.eeg'''</span> in the subdirectory ''Examples/EEG-Focus'' of the BESA Research directory to explain the editing of electrode labels and coordinates: |
| − | + | 1. Select ''File'', then click on ''Open'', or click on <span style="color:#ff9c00;">'''eeg2.eeg '''</span>if this file is contained in the list of currently selected EEG files. | |
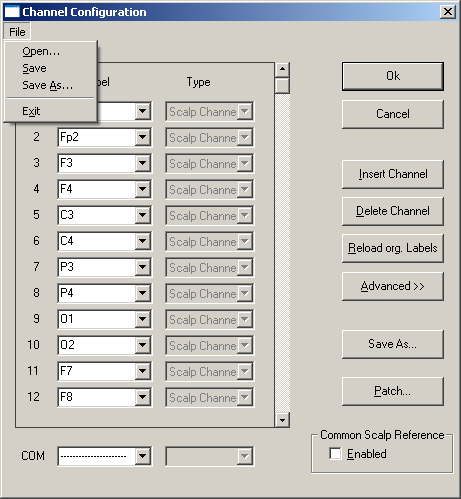
| − | + | 2. Select ''Edit'', then click ''Channel Configuration''. The dialog box shown below will appear. | |
| − | + | ||
| − | + | ||
| − | + | ||
| Line 285: | Line 261: | ||
At the upper left of the figure you see the dropdown menu after selecting the <span style="color:#3366ff;">'''File '''</span>menu in the dialog box. This menu allows to edit a new ('''''New''''') or an existing ('''''Open''''') electrode file and to save the changes to the same ('''''Save''''') or a different ('''''Save As''''') file. Normally, it will not be necessary to use this menu. The control fields on the right will be sufficient. If you type ''''''Ok'''''', you will be given the option of saving the changes to a file. | At the upper left of the figure you see the dropdown menu after selecting the <span style="color:#3366ff;">'''File '''</span>menu in the dialog box. This menu allows to edit a new ('''''New''''') or an existing ('''''Open''''') electrode file and to save the changes to the same ('''''Save''''') or a different ('''''Save As''''') file. Normally, it will not be necessary to use this menu. The control fields on the right will be sufficient. If you type ''''''Ok'''''', you will be given the option of saving the changes to a file. | ||
| − | + | 3. Click on <span style="color:#3366ff;">'''Reload org. </span><span style="color:#3366ff;">Labels'''</span> to reread the original labels as stored in the file header of <span style="color:#ff9c00;">'''eeg2.eeg'''</span>. BESA Research quits editing and redisplays the EEG. Repeat step 2 and select "''Edit / Channel'' ''Configuration''" again. Note: The button <span style="color:#3366ff;">'''Reload org. Labels'''</span> is not available if there are no labels in the file header. | |
| − | + | 4. Click on the empty space of the scroll bar below the <span style="color:#3366ff;">'''scroll '''</span>button and on the <span style="color:#3366ff;">'''down arrow'''</span> of the '''scroll bar''' to display the remaining electrodes in the file. | |
| − | + | 5. Click on electrode '''R''' (line 32), then on the button <span style="color:#3366ff;">'''Delete Electrode'''</span> to remove the associated channel, which does not contain any signal. Note that you may not omit intermediate channels, even if they do not exhibit signals, because the correct correspondence between the series of electrodes and the EEG channels will not be maintained. Use the <span style="color:#3366ff;">'''"Edit / Bad Channels"'''</span> menu to disable artifactual or empty channels. | |
| − | + | 6. Click on''' EOG''' (line 30) and change the entry to '''EOG1'''. Do not type <span style="color:#3366ff;">'''<Enter>,'''</span> but click on the next or a different electrode box to accept the changes. | |
| − | + | 7. '''Double click''' on '''EOG '''(line 31). This will highlight the entry. Simply type the new name '''EOG2''', and note that the old label is replaced when highlighted. Electrodes '''30 '''and '''31 '''are now defined as distinct electrodes. Next, we want to replace the label '''T10 '''by '''A2'''. | |
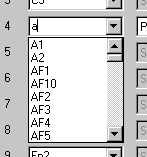
| − | + | 8. Click on the label '''T10''' (line 24). Then click on the <span style="color:#3366ff;">'''drop down'''</span> arrow right of the highlighted label to obtain the list of default scalp electrodes (read from <span style="color:#ff9c00;">'''default.ecd'''</span> and sorted alphabetically). Type '''A''' to jump to the electrodes beginning with letter A (see below). Type '''2''' or '''click''' on '''A2''' in the list. Click on the '''type '''box (Scalp channel) to close the list and display the new entry in line 24. Note that this is the most convenient way to edit an electrode label. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| Line 300: | Line 272: | ||
| − | + | 9. Exercise: repeat step 8 to replace '''T9''' by '''A1'''. Restore labels '''T10''' and '''T9''' in lines 24 and 21. | |
| − | + | 10. Replace SO1 and SO2 (supra-orbital) by '''PSO1 '''and '''PSO2 '''and note that these electrodes are changed to the''' 'Polygraphy'''' type, because no coordinates are associated with these labels. | |
| − | + | 11. After you click <span style="color:#3366ff;">''''OK''''</span>, the box '''Write Channel Configuration File''' will appear and display a name for the current electrode file. By default, the BESA electrode file path and current file name will be used and supplemented by the extension <span style="color:#ff9c00;">''''.elb''''</span>. The electrode file path may be set in the <span style="color:#ff9c00;">'''BESA.ini'''</span> file [Defaults] section under ElectrodeFilePath. If no electrode file path is specified in the <span style="color:#ff9c00;">'''BESA.ini'''</span> file, the default electrode file path ''Montages\Channels ''is used. Simply click '<span style="color:#3366ff;">'''OK'''</span>' or type <span style="color:#3366ff;">'''<Enter> '''</span>to save the changes to this file, or select a new file name and/or path, if you do not want to store the electrode file in the BESA Research electrode file directory. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
Note that by using the default 10/10 labels (see chapter "''Electrode conventions''") you specify that the associated electrode is a scalp electrode. Hence, different labels must be used for polygraphic, intracranial or MEG channels. After you have entered a new non-scalp label, you may select the type of the electrode/channel amongst the different groups ('''Polygraphy, Intracranial, MEG Channel''') from the drop down list in the ''''''Type'''''' box. This will allow for using separate selection and scaling facilities of the channel group control push-buttons at the right of the screen ('''All, Scp, Pgr, Icr, MEG'''). If you have entered a new non-scalp label and select the type '''Scalp Channel''', or if you click on the ''''Advanced>>'''' field, boxes will appear to enter the spherical coordinates (azimuth and latitude) of this electrode (cf. Fig. 6.5). These features may be used to specify non-standard scalp electrodes. Please check the earlier sections of this chapter for electrode conventions. You may view the locations of the scalp electrodes on the head schemes in the mapping window. Select '''Show Electrodes in Maps''' in the "<span style="color:#3366ff;">'''View / Options'''</span>" menu. | Note that by using the default 10/10 labels (see chapter "''Electrode conventions''") you specify that the associated electrode is a scalp electrode. Hence, different labels must be used for polygraphic, intracranial or MEG channels. After you have entered a new non-scalp label, you may select the type of the electrode/channel amongst the different groups ('''Polygraphy, Intracranial, MEG Channel''') from the drop down list in the ''''''Type'''''' box. This will allow for using separate selection and scaling facilities of the channel group control push-buttons at the right of the screen ('''All, Scp, Pgr, Icr, MEG'''). If you have entered a new non-scalp label and select the type '''Scalp Channel''', or if you click on the ''''Advanced>>'''' field, boxes will appear to enter the spherical coordinates (azimuth and latitude) of this electrode (cf. Fig. 6.5). These features may be used to specify non-standard scalp electrodes. Please check the earlier sections of this chapter for electrode conventions. You may view the locations of the scalp electrodes on the head schemes in the mapping window. Select '''Show Electrodes in Maps''' in the "<span style="color:#3366ff;">'''View / Options'''</span>" menu. | ||
| Line 314: | Line 280: | ||
[[Image:ST Electrodes (29).gif]] | [[Image:ST Electrodes (29).gif]] | ||
| − | |||
| − | |||
| Line 324: | Line 288: | ||
'''Edit Common Scalp Reference''' | '''Edit Common Scalp Reference''' | ||
| − | |||
There is a separate line at the bottom in the ''Channel Configuration dialog box'' to enter the label and coordinates of the '''Common Scalp Reference electrode'''. If this is specified and enabled (click on field '''Enabled'''), the information provided by the fact that all scalp electrodes were recorded against a common recording reference will be used for mapping, source imaging and virtual montages. This information will be lost if the common reference has not been specified or if a combination of electrodes has been used as reference during recording. Specify the '''Common Scalp Reference electrode''' only if all electrodes have been referenced to the same single electrode and if a standard 10/10 location has been used for the common recording reference. | There is a separate line at the bottom in the ''Channel Configuration dialog box'' to enter the label and coordinates of the '''Common Scalp Reference electrode'''. If this is specified and enabled (click on field '''Enabled'''), the information provided by the fact that all scalp electrodes were recorded against a common recording reference will be used for mapping, source imaging and virtual montages. This information will be lost if the common reference has not been specified or if a combination of electrodes has been used as reference during recording. Specify the '''Common Scalp Reference electrode''' only if all electrodes have been referenced to the same single electrode and if a standard 10/10 location has been used for the common recording reference. | ||
| − | + | ''Note that BESA Research cannot process digital EEG data correctly if there is no common recording reference'', and if different recording references were used for the various scalp electrodes. For intracranial and polygraphic channels different references may be used. It is preferable to use the common reference also for electrode channels near the eyes, because these electrodes provide valuable information for mapping, source imaging and interpolated montages. The traditional bipolar channels (e.g. horizontal and vertical ''EOG'') may be ''reconstructed digitally'' using the '''Selected Channels''' group or user-defined montages. | |
| + | |||
== 3D Coordinates for Precise Analysis == | == 3D Coordinates for Precise Analysis == | ||
| Line 341: | Line 305: | ||
Assume file name is <span style="color:#ff9c00;">'''datafile.xxx'''</span>. datafile is the base name, <span style="color:#ff9c00;">'''.xxx'''</span> is the extension. Replace the text ''datafile'' by the base name of your own file, and the extension<span style="color:#ff9c00;"><span style="color:#00000a;">'' xxx''</span></span> by the extension of your own file. | Assume file name is <span style="color:#ff9c00;">'''datafile.xxx'''</span>. datafile is the base name, <span style="color:#ff9c00;">'''.xxx'''</span> is the extension. Replace the text ''datafile'' by the base name of your own file, and the extension<span style="color:#ff9c00;"><span style="color:#00000a;">'' xxx''</span></span> by the extension of your own file. | ||
| − | |||
'''Channel definitions for EEG:''' | '''Channel definitions for EEG:''' | ||
| − | |||
Labels have 10-10 names: default locations will be used. | Labels have 10-10 names: default locations will be used. | ||
| Line 359: | Line 321: | ||
'''Define head center.''' No coregistration file exists (<span style="color:#ff9c00;">'''<nowiki>*.sfh</nowiki>'''</span>, see above). File <span style="color:#ff9c00;">'''datafile.cot'''</span> exists, or file <span style="color:#ff9c00;">'''default.cot'''</span> exists, or file <span style="color:#ff9c00;">'''..\default.cot'''</span> exists (i.e. file<span style="color:#ff9c00;">''' default.cot</span><span style="color:#ff9c00;"> '''</span>one folder above the data file): head center as computed by fitting a sphere to the surface points is replaced by the head center coordinates contained in this file. See chapter ''"Working with additional files / cot (Head center) file".'' | '''Define head center.''' No coregistration file exists (<span style="color:#ff9c00;">'''<nowiki>*.sfh</nowiki>'''</span>, see above). File <span style="color:#ff9c00;">'''datafile.cot'''</span> exists, or file <span style="color:#ff9c00;">'''default.cot'''</span> exists, or file <span style="color:#ff9c00;">'''..\default.cot'''</span> exists (i.e. file<span style="color:#ff9c00;">''' default.cot</span><span style="color:#ff9c00;"> '''</span>one folder above the data file): head center as computed by fitting a sphere to the surface points is replaced by the head center coordinates contained in this file. See chapter ''"Working with additional files / cot (Head center) file".'' | ||
| − | |||
=== Data reading rules for MEG === | === Data reading rules for MEG === | ||
| Line 369: | Line 330: | ||
'''Automatic procedure:''' | '''Automatic procedure:''' | ||
| − | |||
Labels have names defined in the files <span style="color:#ff9c00;">'''bti.ecd'''</span> or <span style="color:#ff9c00;">'''nmag.ecd'''</span>. Channels are interpreted as MEG. However, sensor locations and head surface point locations must be defined in additional files as described below. Mapping and source analysis are not possible without one or more of the following additional files. | Labels have names defined in the files <span style="color:#ff9c00;">'''bti.ecd'''</span> or <span style="color:#ff9c00;">'''nmag.ecd'''</span>. Channels are interpreted as MEG. However, sensor locations and head surface point locations must be defined in additional files as described below. Mapping and source analysis are not possible without one or more of the following additional files. | ||
| Line 379: | Line 339: | ||
'''MEG+EEG.''' Define head surface point/electrode coordinates. File <span style="color:#ff9c00;">'''datafile.sfp'''</span>'' ''exists, or file <span style="color:#ff9c00;">'''default.sfp'''</span> exists, or file <span style="color:#ff9c00;">'''..\default.sfp'''</span> exists (i.e. file <span style="color:#ff9c00;">'''default.sfp'''</span> one folder above the data file): electrode coordinates will be replaced/defined by the coordinates defined in this file. If <span style="color:#ff9c00;">'''datafile.sfn'''</span> does not exist, labels can also be defined in this file. See chapter “''Working with additional files/ sfp (surface point'' ''coordinate) file''”. The labels of electrode coordinates '''must '''match to those defined for the data channels. BESA Research will use the labels to associate coordinates with the correct channel. | '''MEG+EEG.''' Define head surface point/electrode coordinates. File <span style="color:#ff9c00;">'''datafile.sfp'''</span>'' ''exists, or file <span style="color:#ff9c00;">'''default.sfp'''</span> exists, or file <span style="color:#ff9c00;">'''..\default.sfp'''</span> exists (i.e. file <span style="color:#ff9c00;">'''default.sfp'''</span> one folder above the data file): electrode coordinates will be replaced/defined by the coordinates defined in this file. If <span style="color:#ff9c00;">'''datafile.sfn'''</span> does not exist, labels can also be defined in this file. See chapter “''Working with additional files/ sfp (surface point'' ''coordinate) file''”. The labels of electrode coordinates '''must '''match to those defined for the data channels. BESA Research will use the labels to associate coordinates with the correct channel. | ||
| − | '''Define sensor coordinates'''. File <span style="color:#ff9c00;">'''datafile.pos'''</span> or <span style="color:#ff9c00;">'''datafile.pmg'''</span> exists, or file <span style="color:#ff9c00;">'''default.pos(.pmg)'''</span> exists, or file <span style="color:#ff9c00;">'''..\default.pos(.pmg)'''</span> exists (i.e. file <span style="color:#ff9c00;">'''default.pos(.pmg)'''</span> one folder above the data file): coordinates are defined in this file. The convention is that'' pos'' files contain gradiometer coordinates and'' pmg'' files contain magnetometer coordinates. This is not necessary for the program to read in values properly: the program makes its decision about the sensor type on the basis of the number of coordinate values | + | '''Define sensor coordinates'''. File <span style="color:#ff9c00;">'''datafile.pos'''</span> or <span style="color:#ff9c00;">'''datafile.pmg'''</span> exists, or file <span style="color:#ff9c00;">'''default.pos(.pmg)'''</span> exists, or file <span style="color:#ff9c00;">'''..\default.pos(.pmg)'''</span> exists (i.e. file <span style="color:#ff9c00;">'''default.pos(.pmg)'''</span> one folder above the data file): coordinates are defined in this file. The convention is that'' pos'' files contain gradiometer coordinates and'' pmg'' files contain magnetometer coordinates. This is not necessary for the program to read in values properly: the program makes its decision about the sensor type on the basis of the number of coordinate values on one line in the file (6 = magnetometers, 9 = gradiometers). See chapter “''Working with additional files/ pos or pmg (MEG sensor coordinate) file”.'' |
| − | + | ||
| − | on one line in the file (6 = magnetometers, 9 = gradiometers). See chapter “''Working with additional files/ pos or pmg (MEG sensor coordinate) file”.'' | + | |
'''Define coregistration information.''' File <span style="color:#ff9c00;">'''datafile.sfh'''</span> exists, or file<span style="color:#ff9c00;">''' default.sfh</span><span style="color:#ff9c00;"> '''</span>exists, or file <span style="color:#ff9c00;">'''..\default.sfh'''</span> exists (i.e. file <span style="color:#ff9c00;">'''default.sfh'''</span> one folder above the data file): head center and relative position of the unit sphere with respect to the head coordinate system is determined by the coregistration of the head coordinates with MRI. See (online) help chapter ''"Integration with MRI and fMRI".'' | '''Define coregistration information.''' File <span style="color:#ff9c00;">'''datafile.sfh'''</span> exists, or file<span style="color:#ff9c00;">''' default.sfh</span><span style="color:#ff9c00;"> '''</span>exists, or file <span style="color:#ff9c00;">'''..\default.sfh'''</span> exists (i.e. file <span style="color:#ff9c00;">'''default.sfh'''</span> one folder above the data file): head center and relative position of the unit sphere with respect to the head coordinate system is determined by the coregistration of the head coordinates with MRI. See (online) help chapter ''"Integration with MRI and fMRI".'' | ||
'''Define head center.''' No coregistration file exists (<span style="color:#ff9c00;">'''<nowiki>*.sfh</nowiki>'''</span>, see above). File <span style="color:#ff9c00;">'''datafile.cot'''</span> exists, or file <span style="color:#ff9c00;">'''default.cot'''</span> exists, or file <span style="color:#ff9c00;">'''..\default.cot'''</span> exists (i.e. file <span style="color:#ff9c00;">'''default.cot</span><span style="color:#ff9c00;"> '''</span>one folder above the data file): head center as computed by fitting a sphere to the surface points is replaced by the head center coordinates contained in this file. See chapter ''"Working with additional files / cot (Head center) file".'' | '''Define head center.''' No coregistration file exists (<span style="color:#ff9c00;">'''<nowiki>*.sfh</nowiki>'''</span>, see above). File <span style="color:#ff9c00;">'''datafile.cot'''</span> exists, or file <span style="color:#ff9c00;">'''default.cot'''</span> exists, or file <span style="color:#ff9c00;">'''..\default.cot'''</span> exists (i.e. file <span style="color:#ff9c00;">'''default.cot</span><span style="color:#ff9c00;"> '''</span>one folder above the data file): head center as computed by fitting a sphere to the surface points is replaced by the head center coordinates contained in this file. See chapter ''"Working with additional files / cot (Head center) file".'' | ||
| + | |||
=== Reading MEG files in ASCII format === | === Reading MEG files in ASCII format === | ||
| − | |||
| − | |||
'''BESA Research uses labeling or channel type definitions to decide whether channels are EEG or MEG. '''Based on the labels defined for the channels, or the type specified by the channel definition file, the program will try to find auxiliary files that define electrode coordinates or MEG sensors. | '''BESA Research uses labeling or channel type definitions to decide whether channels are EEG or MEG. '''Based on the labels defined for the channels, or the type specified by the channel definition file, the program will try to find auxiliary files that define electrode coordinates or MEG sensors. | ||
| − | + | BESA Research uses four files to make its decision: | |
| − | BESA Research uses four files to make its decision:* | + | * <span style="color:#ff9c00;">'''.ela/.elp'''</span> The channel type defined here overrides definitions in <span style="color:#ff9c00;">'''<nowiki>*.ecd</nowiki>'''</span> (below). |
| − | * | + | * <span style="color:#ff9c00;">'''default.ecd'''</span> defines electrode labels and default spherical coordinates based on the 10-20 and 10-10 naming system |
| − | * | + | * <span style="color:#ff9c00;">'''bti.ecd'''</span> defines labels and default spherical coordinates for the BTi whole-head system |
* <span style="color:#ff9c00;">'''nmag.ecd'''</span> defines labels and default spherical coordinates for the Neuromag whole-head system | * <span style="color:#ff9c00;">'''nmag.ecd'''</span> defines labels and default spherical coordinates for the Neuromag whole-head system | ||
| − | |||
| − | |||
| − | |||
| − | |||
If the program finds a label in <span style="color:#ff9c00;">'''default.ecd'''</span>, the channel will automatically be defined as EEG. If not, if it finds a label in one of the other files, the channel will be defined as MEG. If it doesn't find the label anywhere, the channel will be defined as Polygraphic. | If the program finds a label in <span style="color:#ff9c00;">'''default.ecd'''</span>, the channel will automatically be defined as EEG. If not, if it finds a label in one of the other files, the channel will be defined as MEG. If it doesn't find the label anywhere, the channel will be defined as Polygraphic. | ||
The spherical coordinates defined in these files are sufficient for mapping the data. Auxiliary files defining the real sensor coordinates are required for source analysis. | The spherical coordinates defined in these files are sufficient for mapping the data. Auxiliary files defining the real sensor coordinates are required for source analysis. | ||
| − | |||
| − | |||
'''Defining default label and coordinate file for a new MEG system''' | '''Defining default label and coordinate file for a new MEG system''' | ||
| − | |||
When preparing an MEG from a system other than BTi-WHS or Neuromag for import to BESA Research, you should edit either <span style="color:#ff9c00;">'''bti.ecd'''</span> or <span style="color:#ff9c00;">'''nmag.ecd '''</span>to conform with your system. If sensor files (<span style="color:#ff9c00;">'''<nowiki>*.pos/*.pmg</nowiki>'''</span>) are always available for your files, the coordinates in the ''ecd ''files are irrelevant: all you need do is define the labels for your own MEG system or use the labels as already defined. | When preparing an MEG from a system other than BTi-WHS or Neuromag for import to BESA Research, you should edit either <span style="color:#ff9c00;">'''bti.ecd'''</span> or <span style="color:#ff9c00;">'''nmag.ecd '''</span>to conform with your system. If sensor files (<span style="color:#ff9c00;">'''<nowiki>*.pos/*.pmg</nowiki>'''</span>) are always available for your files, the coordinates in the ''ecd ''files are irrelevant: all you need do is define the labels for your own MEG system or use the labels as already defined. | ||
| − | |||
| − | |||
'''Files to prepare for reading in each data file''' | '''Files to prepare for reading in each data file''' | ||
| − | |||
Each auxiliary file should have the same base name as your data file. | Each auxiliary file should have the same base name as your data file. | ||
| − | '''Define channel labels.''' There are several possibilities:* | + | '''Define channel labels.''' There are several possibilities: |
| + | * Generate your data file according to the BESA'' avr'' ''format or the ASCII multiplexed format.'' Labels are listed in the second line of the file. | ||
* Generate a ''label file'' (extension <span style="color:#ff9c00;">'''.ela'''</span>) with one MEG channel label per line (matching with your ''ecd ''file as defined above) or with the type "MEG" and a label for each line. | * Generate a ''label file'' (extension <span style="color:#ff9c00;">'''.ela'''</span>) with one MEG channel label per line (matching with your ''ecd ''file as defined above) or with the type "MEG" and a label for each line. | ||
| − | |||
| − | |||
| − | |||
| − | |||
Label definitions are also possible using ''elp'' or ''elb'' files, but the above two solutions are recommended because they are the simplest. | Label definitions are also possible using ''elp'' or ''elb'' files, but the above two solutions are recommended because they are the simplest. | ||
| Line 437: | Line 381: | ||
'''Define coordinates of the center of the head.''' Generate a ''cot'' file. If this file is absent, BESA Research generates the coordinates by fitting a sphere to the head surface points. | '''Define coordinates of the center of the head.''' Generate a ''cot'' file. If this file is absent, BESA Research generates the coordinates by fitting a sphere to the head surface points. | ||
| − | |||
Note that all coordinates should be within the same frame of reference, i.e. the same coordinate system. Units must be in meters, centimeters, or millimeters. | Note that all coordinates should be within the same frame of reference, i.e. the same coordinate system. Units must be in meters, centimeters, or millimeters. | ||
| + | |||
== Example: Defining Channel Labels == | == Example: Defining Channel Labels == | ||
| − | The files described in these examples can be found in the ''.\Examples\Xtras\EEG+Channel | + | The files described in these examples can be found in the ''.\Examples\Xtras\EEG+Channel Labels'' subdirectory. |
| − | + | ||
| − | + | ||
The simplest way to define electrode coordinates is to use BESA Research’s default settings (defined in the file, <span style="color:#ff9c00;">'''default.ecd'''</span>). In this case, you only need to provide a list of channel labels. If a channel label is defined in <span style="color:#ff9c00;">'''default.ecd'''</span> (i.e. if the labels belong to the 10-20 or 10-10 system), BESA Research will recognize the channel as EEG, and will allocate 3D coordinates. | The simplest way to define electrode coordinates is to use BESA Research’s default settings (defined in the file, <span style="color:#ff9c00;">'''default.ecd'''</span>). In this case, you only need to provide a list of channel labels. If a channel label is defined in <span style="color:#ff9c00;">'''default.ecd'''</span> (i.e. if the labels belong to the 10-20 or 10-10 system), BESA Research will recognize the channel as EEG, and will allocate 3D coordinates. | ||
| − | Labels are not always supplied correctly in the data file. You can override the internal labels in several ways:* Read the data file, and then use "''Edit / Channel Configuration''" to redefine the channels. The configuration is stored in a file with the name <span style="color:#ff9c00;">'''basename.elb'''</span> (for binary data) or <span style="color:#ff9c00;">'''basename.elp'''</span> (for ASCII data), where basename is the base name (name without the extension) of your data file. | + | Labels are not always supplied correctly in the data file. You can override the internal labels in several ways: |
| + | * Read the data file, and then use "''Edit / Channel Configuration''" to redefine the channels. The configuration is stored in a file with the name <span style="color:#ff9c00;">'''basename.elb'''</span> (for binary data) or <span style="color:#ff9c00;">'''basename.elp'''</span> (for ASCII data), where basename is the base name (name without the extension) of your data file. | ||
* Prepare a label file (with the extension ''ela'') containing a list of labels. This can also specify channel types (e.g. EEG, Polygraphic, Intracranial, MEG). | * Prepare a label file (with the extension ''ela'') containing a list of labels. This can also specify channel types (e.g. EEG, Polygraphic, Intracranial, MEG). | ||
* Prepare a file (with the extension ''elp'') containing spherical coordinates of the channels. This is the method used with the previous version of BESA Research. If the file doesn’t contain labels, labels are allocated based on their proximity to the 10-20 or 10-10 definitions in <span style="color:#ff9c00;">'''default.ecd'''</span>. | * Prepare a file (with the extension ''elp'') containing spherical coordinates of the channels. This is the method used with the previous version of BESA Research. If the file doesn’t contain labels, labels are allocated based on their proximity to the 10-20 or 10-10 definitions in <span style="color:#ff9c00;">'''default.ecd'''</span>. | ||
| − | |||
| − | |||
| − | |||
| − | |||
The following examples illustrate the above three methods. The input files are all in ''BESA avr format, ''although these examples apply to all EEG data formats in which only EEG channels exist. | The following examples illustrate the above three methods. The input files are all in ''BESA avr format, ''although these examples apply to all EEG data formats in which only EEG channels exist. | ||
| Line 462: | Line 401: | ||
| − | '''Example 1. EEG file containing wrong labels – use ''Edit/Channel Configuration ''to redefine labels'''* The'' avr'' file, <span style="color:#ff9c00;">'''EEGwithLabels.avr'''</span>, contains the EEG labels. Channels 4 and 14 have been mislabeled – the labels need to be swapped. | + | '''Example 1. EEG file containing wrong labels – use ''Edit/Channel Configuration ''to redefine labels''' |
| + | * The'' avr'' file, <span style="color:#ff9c00;">'''EEGwithLabels.avr'''</span>, contains the EEG labels. Channels 4 and 14 have been mislabeled – the labels need to be swapped. | ||
* Open the file with '''''File/Open''''' (Select file type ''BESA avr''. Find the correct directory ''Xtras\EEG+Channel Labels''). | * Open the file with '''''File/Open''''' (Select file type ''BESA avr''. Find the correct directory ''Xtras\EEG+Channel Labels''). | ||
* The file should open correctly, displaying 32 channels of EEG. | * The file should open correctly, displaying 32 channels of EEG. | ||
| Line 473: | Line 413: | ||
| − | + | '''Example 2. EEG file with no labels – channel labels in auxiliary file''' | |
| − | + | * The ''avr'' file, <span style="color:#ff9c00;">'''EEGwithoutLabels.avr'''</span>, has no labels. | |
| − | + | ||
| − | '''Example 2. EEG file with no labels – channel labels in auxiliary file'''* The ''avr'' file, <span style="color:#ff9c00;">'''EEGwithoutLabels.avr'''</span>, has no labels. | + | |
* EEG labels are defined in <span style="color:#ff9c00;">'''EEGwithoutLabels.ela'''</span>. This is read automatically when the file is opened. | * EEG labels are defined in <span style="color:#ff9c00;">'''EEGwithoutLabels.ela'''</span>. This is read automatically when the file is opened. | ||
* In this example, labels are correct. Each label in the ''ela'' file is on one line: | * In this example, labels are correct. Each label in the ''ela'' file is on one line: | ||
| − | |||
| − | |||
<div style="margin-left:0cm;margin-right:0cm;">''Fp1''</div> | <div style="margin-left:0cm;margin-right:0cm;">''Fp1''</div> | ||
| Line 495: | Line 431: | ||
| − | '''Example 3. EEG file with no labels – channel labels derived from spherical coordinates'''* The ''avr'' file, <span style="color:#ff9c00;">'''EEGwithSphericalCoords.avr'''</span>, has no labels. | + | '''Example 3. EEG file with no labels – channel labels derived from spherical coordinates''' |
| + | * The ''avr'' file, <span style="color:#ff9c00;">'''EEGwithSphericalCoords.avr'''</span>, has no labels. | ||
* Spherical coordinates are defined in the ''elp'' file, <span style="color:#ff9c00;">'''EEGwithSphericalCoords.elp'''</span>. This contains spherical coordinates (theta and phi) and no labels: | * Spherical coordinates are defined in the ''elp'' file, <span style="color:#ff9c00;">'''EEGwithSphericalCoords.elp'''</span>. This contains spherical coordinates (theta and phi) and no labels: | ||
| − | |||
| − | |||
<div style="margin-left:0cm;margin-right:0cm;">''-93 -72''</div> | <div style="margin-left:0cm;margin-right:0cm;">''-93 -72''</div> | ||
| Line 512: | Line 447: | ||
<div style="margin-left:0cm;margin-right:0cm;">''62 51''</div> | <div style="margin-left:0cm;margin-right:0cm;">''62 51''</div> | ||
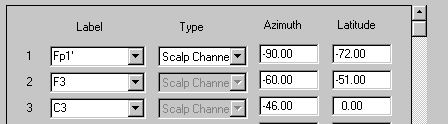
| − | + | * When the data file is opened, the ''elp'' file is read automatically, and BESA Research uses the tables in <span style="color:#ff9c00;">'''default.ecd'''</span> to assign channel labels. To indicate that it has assigned user defined coordinates and matched with the closest standard electrode, BESA appends an apostrophe (‘) to each label: | |
| − | + | ||
| − | + | ||
<div style="margin-left:0cm;margin-right:0cm;">''Fp1’''</div> | <div style="margin-left:0cm;margin-right:0cm;">''Fp1’''</div> | ||
| Line 526: | Line 459: | ||
<div style="margin-left:0cm;margin-right:0cm;">''Fz’''</div> | <div style="margin-left:0cm;margin-right:0cm;">''Fz’''</div> | ||
| − | + | * We advise to assign specific labels as well as spherical coordinates if you want to use your own spherical coordinate system, e.g.: | |
| − | + | ||
| − | + | ||
<div style="margin-left:0cm;margin-right:0cm;">''FP1u -90 -72''</div> | <div style="margin-left:0cm;margin-right:0cm;">''FP1u -90 -72''</div> | ||
| − | + | '''Example 4. EEG file with no labels – channel labels not in basename.el?''' | |
| − | + | * The ''avr ''file, <span style="color:#ff9c00;">'''EEGnoLabelsNoElaFile.avr'''</span>, has no corresponding ''ela, elp'', or ''elb'' file, i.e. no file with the same base name and the ''el?'' extension. | |
| − | '''Example 4. EEG file with no labels – channel labels not in basename.el?'''* | + | |
* When you open the file, BESA Research will ask for a channel configuration file. The ''File Open'' ''dialog ''will select the ''directory .\Montages\Channels''. The idea is that standard (= frequently used) electrode configurations should be kept in this directory. | * When you open the file, BESA Research will ask for a channel configuration file. The ''File Open'' ''dialog ''will select the ''directory .\Montages\Channels''. The idea is that standard (= frequently used) electrode configurations should be kept in this directory. | ||
* Select the file <span style="color:#ff9c00;">'''XtrasExample.ela'''</span>. | * Select the file <span style="color:#ff9c00;">'''XtrasExample.ela'''</span>. | ||
* Close the data file and reopen it. The file will open with the correct labels. In the BESA window title you will see that the file <span style="color:#ff9c00;">'''EEGnoLabelsNoElaFile.elp'''</span> has been read automatically. This file was created when <span style="color:#ff9c00;">'''XtrasExample.ela'''</span> was read. | * Close the data file and reopen it. The file will open with the correct labels. In the BESA window title you will see that the file <span style="color:#ff9c00;">'''EEGnoLabelsNoElaFile.elp'''</span> has been read automatically. This file was created when <span style="color:#ff9c00;">'''XtrasExample.ela'''</span> was read. | ||
| − | |||
| Line 557: | Line 486: | ||
<div style="margin-left:0cm;margin-right:0cm;">''F3''</div> | <div style="margin-left:0cm;margin-right:0cm;">''F3''</div> | ||
| − | |||
The prefix "''POLY''" specifies that the channel is polygraphic. Most other channels are interpreted as EEG because the labels are known in the 10-20 system. | The prefix "''POLY''" specifies that the channel is polygraphic. Most other channels are interpreted as EEG because the labels are known in the 10-20 system. | ||
| Line 564: | Line 492: | ||
Note that you can also define channels as EEG by specifying the ''"EEG" ''prefix (e.g. ''"EEG E1". ''This is useful if there are many more channels than are defined in the 10-10 or 10-20 systems, and if the channel coordinates are defined. | Note that you can also define channels as EEG by specifying the ''"EEG" ''prefix (e.g. ''"EEG E1". ''This is useful if there are many more channels than are defined in the 10-10 or 10-20 systems, and if the channel coordinates are defined. | ||
| + | |||
== Example: EEG with Digitized Coordinates == | == Example: EEG with Digitized Coordinates == | ||
| Line 571: | Line 500: | ||
In the previous examples, we have illustrated how to assign labels to channels using channel definition files. In those examples, only spherical coordinates were defined. Here we will show how to read digitized surface points into BESA Research, using the surface point (''sfp'') coordinate file and the surface point name (''sfn'') file. | In the previous examples, we have illustrated how to assign labels to channels using channel definition files. In those examples, only spherical coordinates were defined. Here we will show how to read digitized surface points into BESA Research, using the surface point (''sfp'') coordinate file and the surface point name (''sfn'') file. | ||
| − | The principles of defining digitization coordinate files are:* The labels in the ''sfp/sfn'' file combination are used to assign coordinates to electrodes. Thus, if a coordinate has the name ‘''Fz''’ it will be assigned to the channel with the label ‘''Fz''’. | + | The principles of defining digitization coordinate files are: |
| + | * The labels in the ''sfp/sfn'' file combination are used to assign coordinates to electrodes. Thus, if a coordinate has the name ‘''Fz''’ it will be assigned to the channel with the label ‘''Fz''’. | ||
* In consequence, digitization of surface points can be in a different order to the sequence of channels in the data file. Matching to channels is done by comparing the labels. | * In consequence, digitization of surface points can be in a different order to the sequence of channels in the data file. Matching to channels is done by comparing the labels. | ||
* We recommend that the fiducial points, '''nasion, left preauricular point, right preauricular point''' be digitized. If you do not digitize them, BESA Research will simulate these locations (see ''“Example: Digitization points with and without Fiducials”''). Fiducial points, labeled '''FidNz, FidT9, FidT10''' should be the first three coordinates in the ''sfp'' file. | * We recommend that the fiducial points, '''nasion, left preauricular point, right preauricular point''' be digitized. If you do not digitize them, BESA Research will simulate these locations (see ''“Example: Digitization points with and without Fiducials”''). Fiducial points, labeled '''FidNz, FidT9, FidT10''' should be the first three coordinates in the ''sfp'' file. | ||
* As with the channel definition files, it is easiest for BESA Research if you name the ''sfp/sfn'' files using the base name of the data file, e.g. if the data file is named <span style="color:#ff9c00;">'''doodah.avr'''</span>, name the'' sfp'' file <span style="color:#ff9c00;">'''doodah.sfp'''</span> and the ''sfn'' file <span style="color:#ff9c00;">'''doodah.sfn'''</span>. | * As with the channel definition files, it is easiest for BESA Research if you name the ''sfp/sfn'' files using the base name of the data file, e.g. if the data file is named <span style="color:#ff9c00;">'''doodah.avr'''</span>, name the'' sfp'' file <span style="color:#ff9c00;">'''doodah.sfp'''</span> and the ''sfn'' file <span style="color:#ff9c00;">'''doodah.sfn'''</span>. | ||
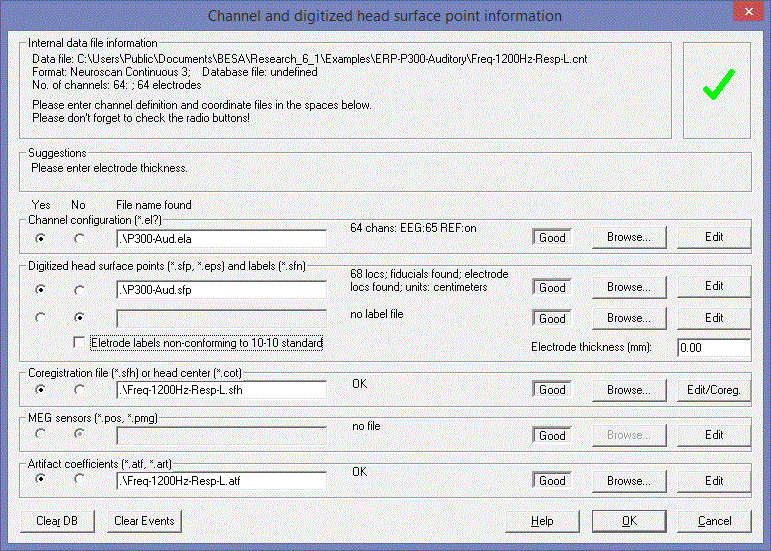
* You specify the files to be read in the ''Channel and digitized head surface point information dialog box.'' | * You specify the files to be read in the ''Channel and digitized head surface point information dialog box.'' | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| Line 586: | Line 511: | ||
| − | '''Example 1. EEG file containing labels, ''sfp'' file containing coordinates, ''sfn ''file containing coordinate names'''* The ''avr'' file, <span style="color:#ff9c00;">'''EEGdigitized1.avr'''</span>, contains the channel labels. Therefore, we don’t need a channel definition file. | + | '''Example 1. EEG file containing labels, ''sfp'' file containing coordinates, ''sfn ''file containing coordinate names''' |
| + | * The ''avr'' file, <span style="color:#ff9c00;">'''EEGdigitized1.avr'''</span>, contains the channel labels. Therefore, we don’t need a channel definition file. | ||
* The ''sfp'' file, <span style="color:#ff9c00;">'''EEGdigitized1.avr'''</span>, contains digitized coordinates of electrodes and of additional surface points. The labels in the file do not correspond to the electrode labels in the ''avr ''file. | * The ''sfp'' file, <span style="color:#ff9c00;">'''EEGdigitized1.avr'''</span>, contains digitized coordinates of electrodes and of additional surface points. The labels in the file do not correspond to the electrode labels in the ''avr ''file. | ||
* The ''sfn'' file contains the corrected labels (1 line for each corresponding line in the'' sfp'' file). Now it is possible to match up electrode labels with the labels in the ''avr ''file. | * The ''sfn'' file contains the corrected labels (1 line for each corresponding line in the'' sfp'' file). Now it is possible to match up electrode labels with the labels in the ''avr ''file. | ||
| Line 594: | Line 520: | ||
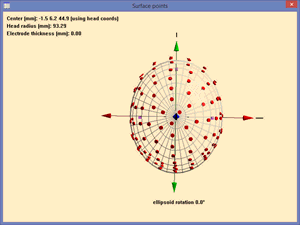
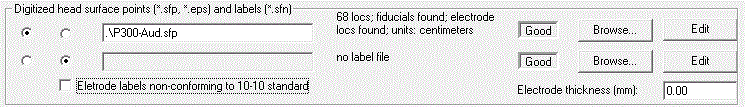
* Define the electrode thickness as 6 mm (at the right of the ‘''Digitized head surface points’'' box. This is the distance of the digitized point on the electrode to the surface of the head. | * Define the electrode thickness as 6 mm (at the right of the ‘''Digitized head surface points’'' box. This is the distance of the digitized point on the electrode to the surface of the head. | ||
* Press <span style="color:#3366ff;">'''‘OK’'''</span> in the dialog box and view the coordinates by pressing the <span style="color:#3366ff;">'''‘V’'''</span> key. | * Press <span style="color:#3366ff;">'''‘OK’'''</span> in the dialog box and view the coordinates by pressing the <span style="color:#3366ff;">'''‘V’'''</span> key. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
'''Example 2. EEG file without labels, channel labels in ''ela'' file, surface point coordinates and names in ''sfp'' file''' | '''Example 2. EEG file without labels, channel labels in ''ela'' file, surface point coordinates and names in ''sfp'' file''' | ||
| − | |||
| − | |||
* The ''avr'' file, <span style="color:#ff9c00;">'''EEGdigitized2.avr'''</span>, has no channel label. Therefore, a label file is required. Here, the label file is <span style="color:#ff9c00;">'''EEGdigitized2.ela'''</span>. | * The ''avr'' file, <span style="color:#ff9c00;">'''EEGdigitized2.avr'''</span>, has no channel label. Therefore, a label file is required. Here, the label file is <span style="color:#ff9c00;">'''EEGdigitized2.ela'''</span>. | ||
* The ''sfp'' file, <span style="color:#ff9c00;">'''EEGdigitized2.sfp'''</span>, contains digitized coordinates of electrodes and of additional surface points. The labels are defined correctly in the ''sfp ''file, i.e. for every EEG channel label there is a corresponding coordinate. Therefore, no ''sfn'' file is required. | * The ''sfp'' file, <span style="color:#ff9c00;">'''EEGdigitized2.sfp'''</span>, contains digitized coordinates of electrodes and of additional surface points. The labels are defined correctly in the ''sfp ''file, i.e. for every EEG channel label there is a corresponding coordinate. Therefore, no ''sfn'' file is required. | ||
* When you open the file, don’t forget to define the electrode thickness as 6 mm in the ''Channel and digitized head surface point information dialog box.'' | * When you open the file, don’t forget to define the electrode thickness as 6 mm in the ''Channel and digitized head surface point information dialog box.'' | ||
| − | |||
| Line 616: | Line 534: | ||
Data from the Polhemus (other digitizers too) may often not fit the format BESA Research requires for the surface point file. Note that Polhemus data can be exported directly into BESA-compatible ''sfp''-files using the LOCATOR software. | Data from the Polhemus (other digitizers too) may often not fit the format BESA Research requires for the surface point file. Note that Polhemus data can be exported directly into BESA-compatible ''sfp''-files using the LOCATOR software. | ||
| − | BESA Research requires either* just the cartesian coordinates (x, y, z) values -- one set of coordinates per line. In this case, labels must be defined in a parallel surface point name file, e.g. | + | BESA Research requires either |
| − | + | * just the cartesian coordinates (x, y, z) values -- one set of coordinates per line. In this case, labels must be defined in a parallel surface point name file, e.g. | |
| − | + | ||
<div style="margin-left:1.27cm;margin-right:0cm;">''0.5 3.75 12.68''</div> | <div style="margin-left:1.27cm;margin-right:0cm;">''0.5 3.75 12.68''</div> | ||
<div style="margin-left:0cm;margin-right:0cm;">or</div> | <div style="margin-left:0cm;margin-right:0cm;">or</div> | ||
| − | |||
* the cartesian coordinates plus a label. The label can be in front of or behind the coordinates on the line, e.g. | * the cartesian coordinates plus a label. The label can be in front of or behind the coordinates on the line, e.g. | ||
| − | |||
| − | |||
<div style="margin-left:1.27cm;margin-right:0cm;">''0.5 3.75 12.68 Fz''</div> | <div style="margin-left:1.27cm;margin-right:0cm;">''0.5 3.75 12.68 Fz''</div> | ||
| Line 634: | Line 548: | ||
<div style="margin-left:1.27cm;margin-right:0cm;">''Fz 0.5 3.75 12.68'' </div> | <div style="margin-left:1.27cm;margin-right:0cm;">''Fz 0.5 3.75 12.68'' </div> | ||
| − | |||
Here is an example of a few lines of a (''sfp'') file that are not read correctly by BESA Research: | Here is an example of a few lines of a (''sfp'') file that are not read correctly by BESA Research: | ||
| Line 651: | Line 564: | ||
<div style="margin-left:0cm;margin-right:0cm;">''4 -60.701 79.631 78.273''</div> | <div style="margin-left:0cm;margin-right:0cm;">''4 -60.701 79.631 78.273''</div> | ||
| − | |||
What is wrong? | What is wrong? | ||
| − | |||
* First, some of the points are just numbered. These numbers don't tell BESA Research which electrode channel to which the coordinates should be assigned – assignments should be via channel labels and not numbers. | * First, some of the points are just numbered. These numbers don't tell BESA Research which electrode channel to which the coordinates should be assigned – assignments should be via channel labels and not numbers. | ||
* Second, Nz, T9, T10 define the fiducials. Instead, the labels FidNz, FidT9, FidT10 are required (prefix "Fid"). | * Second, Nz, T9, T10 define the fiducials. Instead, the labels FidNz, FidT9, FidT10 are required (prefix "Fid"). | ||
| − | |||
| − | |||
| − | |||
| − | |||
What should be done? Probably the best way is | What should be done? Probably the best way is | ||
| − | |||
a) keep only the coordinates in the <span style="color:#ff9c00;">'''<nowiki>*.sfp </nowiki>'''</span>file: | a) keep only the coordinates in the <span style="color:#ff9c00;">'''<nowiki>*.sfp </nowiki>'''</span>file: | ||
| Line 683: | Line 589: | ||
b) prepare a surface point name file (<span style="color:#ff9c00;">'''<nowiki>*.sfn</nowiki>'''</span>) containing the corresponding labels: | b) prepare a surface point name file (<span style="color:#ff9c00;">'''<nowiki>*.sfn</nowiki>'''</span>) containing the corresponding labels: | ||
| − | |||
<div style="margin-left:0cm;margin-right:0cm;">''FidNz''</div> | <div style="margin-left:0cm;margin-right:0cm;">''FidNz''</div> | ||
| Line 698: | Line 603: | ||
<div style="margin-left:0cm;margin-right:0cm;">''F3''</div> | <div style="margin-left:0cm;margin-right:0cm;">''F3''</div> | ||
| − | |||
Keeping labels and coordinates separate means that the label file needs generating only once. The coordinate file is different for each subject. | Keeping labels and coordinates separate means that the label file needs generating only once. The coordinate file is different for each subject. | ||
Alternatively, if your digitizer program attaches the labels correctly to the coordinates, then you can prepare the ''sfp'' file like this: | Alternatively, if your digitizer program attaches the labels correctly to the coordinates, then you can prepare the ''sfp'' file like this: | ||
| − | |||
<div style="margin-left:0cm;margin-right:0cm;">''FidNz 0 87.721 0''</div> | <div style="margin-left:0cm;margin-right:0cm;">''FidNz 0 87.721 0''</div> | ||
| Line 718: | Line 621: | ||
<div style="margin-left:0cm;margin-right:0cm;">''F3 -60.701 79.631 78.273''</div> | <div style="margin-left:0cm;margin-right:0cm;">''F3 -60.701 79.631 78.273''</div> | ||
| + | |||
== Example: Digitization points with and without Fiducials == | == Example: Digitization points with and without Fiducials == | ||
| Line 730: | Line 634: | ||
'''Consequences of omitting fiducials''' | '''Consequences of omitting fiducials''' | ||
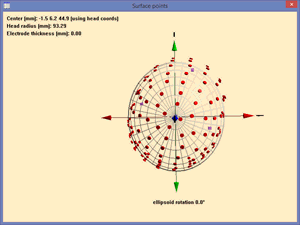
| − | When these files have been read into BESA Research, look at the head surface points in 3D using ''File/Head Surface Points'' ''and Sensors/View'' (or use the shortcut <span style="color:#3366ff;">'''‘V’'''</span>). You will see small differences in fiducial locations between the real and the simulated locations. You can expect very slight influences on the results of source modeling (the spherical head may be rotated slightly, although the head center and | + | When these files have been read into BESA Research, look at the head surface points in 3D using ''File/Head Surface Points'' ''and Sensors/View'' (or use the shortcut <span style="color:#3366ff;">'''‘V’'''</span>). You will see small differences in fiducial locations between the real and the simulated locations. You can expect very slight influences on the results of source modeling (the spherical head may be rotated slightly, although the head center and radius will be identical), and output of source locations in head coordinates will be different, because these coordinates are based on fiducial locations (see chapter ''“Working with Electrodes and Surface'' ''Locations/ Coordinate systems''”). |
| − | |||
== Example: ASCII Import == | == Example: ASCII Import == | ||
| Line 741: | Line 644: | ||
The array can be '''vectorized '''(one channel, all time points, per line) or '''multiplexed''' (one time point, all channels, per line). These alternatives are illustrated in the two example files <span style="color:#ff9c00;">'''vectorized.asc'''</span> and <span style="color:#ff9c00;">'''multiplexed.asc'''</span>, and in the tables below: | The array can be '''vectorized '''(one channel, all time points, per line) or '''multiplexed''' (one time point, all channels, per line). These alternatives are illustrated in the two example files <span style="color:#ff9c00;">'''vectorized.asc'''</span> and <span style="color:#ff9c00;">'''multiplexed.asc'''</span>, and in the tables below: | ||
| − | |||
| − | |||
'''Vectorized array:''' | '''Vectorized array:''' | ||
| − | |||
| − | |||
{| style="border-spacing:0;width:12.993cm;" | {| style="border-spacing:0;width:12.993cm;" | ||
| Line 775: | Line 674: | ||
'''Multiplexed array:''' | '''Multiplexed array:''' | ||
| − | |||
| − | |||
{| style="border-spacing:0;width:12.993cm;" | {| style="border-spacing:0;width:12.993cm;" | ||
| Line 808: | Line 705: | ||
| − | '''Example 1. Vectorized data'''* The file <span style="color:#ff9c00;">'''vectorized.asc'''</span> contains the data. The file should be imported via ''File/Import ASCII File''. | + | '''Example 1. Vectorized data''' |
| + | * The file <span style="color:#ff9c00;">'''vectorized.asc'''</span> contains the data. The file should be imported via ''File/Import ASCII File''. | ||
* First you will be asked for a name for the binary target file. The name <span style="color:#ff9c00;">'''vectorized.fsg'''</span> is suggested. You may accept this name by pressing <span style="color:#3366ff;">'''‘OK’'''</span> or choose an alternative name. Note that if the file already exists, the imported data will be appended to the file. | * First you will be asked for a name for the binary target file. The name <span style="color:#ff9c00;">'''vectorized.fsg'''</span> is suggested. You may accept this name by pressing <span style="color:#3366ff;">'''‘OK’'''</span> or choose an alternative name. Note that if the file already exists, the imported data will be appended to the file. | ||
* Next, the ''ASCII File Properties dialog box'' will open. First select ''‘Vectorized’'', and make sure the subsequent entries are correct: | * Next, the ''ASCII File Properties dialog box'' will open. First select ''‘Vectorized’'', and make sure the subsequent entries are correct: | ||
| − | |||
| − | |||
<div style="margin-left:1.27cm;margin-right:0cm;">Header Lines = 0 (i.e. in this example the numbers start on the first line)</div> | <div style="margin-left:1.27cm;margin-right:0cm;">Header Lines = 0 (i.e. in this example the numbers start on the first line)</div> | ||
| Line 827: | Line 723: | ||
<div style="margin-left:0cm;margin-right:0cm;">Press <span style="color:#3366ff;">'''‘OK’'''</span> to accept the settings.</div> | <div style="margin-left:0cm;margin-right:0cm;">Press <span style="color:#3366ff;">'''‘OK’'''</span> to accept the settings.</div> | ||
| − | |||
* Next, the ''Channel and digitized head surface point information dialog box'' will open. In the ''‘Channel configuration’'' box, the label file, <span style="color:#ff9c00;">'''vectorized.ela'''</span>, will be detected automatically. Automatic detection occurs when the label file has the same base name as the data file (in this case, vectorized). To the right of the file name is a summary of channel types: 32 channels found, 30 are EEG, 1 is intercranial, 1 is polygraphic. | * Next, the ''Channel and digitized head surface point information dialog box'' will open. In the ''‘Channel configuration’'' box, the label file, <span style="color:#ff9c00;">'''vectorized.ela'''</span>, will be detected automatically. Automatic detection occurs when the label file has the same base name as the data file (in this case, vectorized). To the right of the file name is a summary of channel types: 32 channels found, 30 are EEG, 1 is intercranial, 1 is polygraphic. | ||
| − | |||
| − | |||
| − | |||
Note the green tick at the top left of the dialog box. This indicates that BESA Research thinks that it has sufficient information to read the file, and map and do source analysis on the data. | Note the green tick at the top left of the dialog box. This indicates that BESA Research thinks that it has sufficient information to read the file, and map and do source analysis on the data. | ||
| − | |||
* To see how channel types are specified in <span style="color:#ff9c00;">'''vectorized.ela'''</span>, click on the <span style="color:#3366ff;">'''Edit '''</span>button to view the file with the Notepad program. Here you will see that most channels have 10-20 electrode names. Channel 3 has the prefix ‘''POLY''’, specifying that this channel is polygraphic. Channel 31 has the prefix ‘''ICR''’, specifying that this channel is intercranial. Close Notepad, and click ‘<span style="color:#3366ff;">'''OK'''</span>’ in the dialog box. | * To see how channel types are specified in <span style="color:#ff9c00;">'''vectorized.ela'''</span>, click on the <span style="color:#3366ff;">'''Edit '''</span>button to view the file with the Notepad program. Here you will see that most channels have 10-20 electrode names. Channel 3 has the prefix ‘''POLY''’, specifying that this channel is polygraphic. Channel 31 has the prefix ‘''ICR''’, specifying that this channel is intercranial. Close Notepad, and click ‘<span style="color:#3366ff;">'''OK'''</span>’ in the dialog box. | ||
| Line 842: | Line 733: | ||
| + | '''Example 2. Multiplexed data''' | ||
| + | * This example is similar to Example 1. In this case, import the file <span style="color:#ff9c00;">'''multiplexed.asc'''</span>, and select ‘Multiplexed’ in the ''ASCII File Properties dialog box''. Other settings in the dialog box stay as they were. | ||
| − | + | '''Notes''' | |
| − | + | # The numbers in the source files can be split into several lines per channel or per time point. Then you will have to enter the correct number of time points and channels in the dialog box. In the present examples, the lines are not split (the vectorized file has all 640 time points in each line, and the multiplexed file has all 32 channels in each line). In this case, BESA Research selects the correct numbers of time points and channels automatically. | |
| − | ''' | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
# If you have digitized coordinates, these can be specified in the Channel and digitized head surface point information dialog box. Since the procedure is the same as when reading data, this is described elsewhere under “''Example: EEG with Digitized Coordinates''”. | # If you have digitized coordinates, these can be specified in the Channel and digitized head surface point information dialog box. Since the procedure is the same as when reading data, this is described elsewhere under “''Example: EEG with Digitized Coordinates''”. | ||
| − | |||
| Line 863: | Line 748: | ||
'''Multiplexed MEG ASCII file with labels in the header (<span style="color:#ff9c00;"><span style="color:#00000a;">''med.mul''</span></span>)''' | '''Multiplexed MEG ASCII file with labels in the header (<span style="color:#ff9c00;"><span style="color:#00000a;">''med.mul''</span></span>)''' | ||
| − | + | For reading MEG data, BESA Research expects | |
| − | For reading MEG data, BESA Research expects* Correct channel definitions, i.e. channels should be defined as MEG. | + | * Correct channel definitions, i.e. channels should be defined as MEG. |
* Head surface points. | * Head surface points. | ||
* Sensor coordinates, in '''the same coordinate system''' as the head surface points. | * Sensor coordinates, in '''the same coordinate system''' as the head surface points. | ||
* Optionally, you can define the coordinates of the center of the head. This will be important if too few head surface points are available to specify where to place the spherical head used by BESA Research for source modeling, or if you want to use some external definition, e.g. from the MRI. | * Optionally, you can define the coordinates of the center of the head. This will be important if too few head surface points are available to specify where to place the spherical head used by BESA Research for source modeling, or if you want to use some external definition, e.g. from the MRI. | ||
| − | |||
| − | |||
| − | |||
| − | |||
As with digitized EEG coordinates, we use the ''Channel and digitized head surface point information'' ''dialog box'' to specify the files which need to be read. | As with digitized EEG coordinates, we use the ''Channel and digitized head surface point information'' ''dialog box'' to specify the files which need to be read. | ||
| Line 877: | Line 758: | ||
'''Example 1. File Open''' | '''Example 1. File Open''' | ||
| − | |||
| − | |||
* The file, <span style="color:#ff9c00;">'''MEGASCII.mul'''</span>, contains MEG data in the ASCII multiplexed format. This format contains channel labels. The labels used are recognized by BESA Research as originating from the Neuromag system. They are therefore identified as MEG and do not need further identification. | * The file, <span style="color:#ff9c00;">'''MEGASCII.mul'''</span>, contains MEG data in the ASCII multiplexed format. This format contains channel labels. The labels used are recognized by BESA Research as originating from the Neuromag system. They are therefore identified as MEG and do not need further identification. | ||
* The ''sfp'' file, <span style="color:#ff9c00;">'''MEGASCII.sfp'''</span>, defines fiducials and head surface points. Coordinate labels are included in the file, so no'' sfn'' file is required. | * The ''sfp'' file, <span style="color:#ff9c00;">'''MEGASCII.sfp'''</span>, defines fiducials and head surface points. Coordinate labels are included in the file, so no'' sfn'' file is required. | ||
| Line 885: | Line 764: | ||
* Open the file, selecting current file type as ''‘*,m''??’. The ''Channel and digitized head surface point'' ''information dialog box'' will open, displaying the different auxiliary file names. The green tick indicates that BESA Research finds everything to be OK. | * Open the file, selecting current file type as ''‘*,m''??’. The ''Channel and digitized head surface point'' ''information dialog box'' will open, displaying the different auxiliary file names. The green tick indicates that BESA Research finds everything to be OK. | ||
* Press the <span style="color:#3366ff;">'''‘OK’'''</span> button and then the<span style="color:#3366ff;">''' ‘V’ '''</span>key to view the coordinates. | * Press the <span style="color:#3366ff;">'''‘OK’'''</span> button and then the<span style="color:#3366ff;">''' ‘V’ '''</span>key to view the coordinates. | ||
| − | |||
| − | |||
| − | |||
'''Example 2. File Import''' | '''Example 2. File Import''' | ||
| − | |||
| − | |||
* The file, <span style="color:#ff9c00;">'''MEGASCIIimport.asc'''</span>, contains MEG data in a multiplexed array, without a header. This needs to be imported using ''File/Import ASCII'' (see ''“Example: ASCII Import”).'' | * The file, <span style="color:#ff9c00;">'''MEGASCIIimport.asc'''</span>, contains MEG data in a multiplexed array, without a header. This needs to be imported using ''File/Import ASCII'' (see ''“Example: ASCII Import”).'' | ||
* On import you have to specify the file as ‘Multiplexed’, the number of time points (285), the number of channels (132), the bins/µV (or bins/fT) (=1), the time at which the stimulus occurred (50 ms), and the sampling rate (949.667 Hz). | * On import you have to specify the file as ‘Multiplexed’, the number of time points (285), the number of channels (132), the bins/µV (or bins/fT) (=1), the time at which the stimulus occurred (50 ms), and the sampling rate (949.667 Hz). | ||
| Line 898: | Line 772: | ||
* Since it recognizes the channels as MEG, the ''Channel and digitized head surface point information dialog box'' will open, displaying the different auxiliary file names as before. Since all necessary files with the same base name as the data file are supplied, they are read automatically. | * Since it recognizes the channels as MEG, the ''Channel and digitized head surface point information dialog box'' will open, displaying the different auxiliary file names as before. Since all necessary files with the same base name as the data file are supplied, they are read automatically. | ||
* Press the ‘<span style="color:#3366ff;">'''OK’'''</span> button, enter a segment name, and then the <span style="color:#3366ff;">'''‘V’'''</span> key to view the coordinates. | * Press the ‘<span style="color:#3366ff;">'''OK’'''</span> button, enter a segment name, and then the <span style="color:#3366ff;">'''‘V’'''</span> key to view the coordinates. | ||
| − | |||
| − | |||
| − | |||
'''Example 3. File Open -- MEG information recorded elsewhere''' | '''Example 3. File Open -- MEG information recorded elsewhere''' | ||
| + | This example illustrates the case where the auxiliary files have a different base name from the data file: you must select the file name in the ''Channel and digitized head surface point information dialog box''. | ||
| − | + | * Open the file, <span style="color:#ff9c00;">'''MEGASCIIelsewhere.mul'''</span>. It is read as an MEG magnetometer file. | |
* In the ''Channel and digitized head surface point information dialog box'', specify | * In the ''Channel and digitized head surface point information dialog box'', specify | ||
| − | |||
| − | |||
| − | |||
<div style="margin-left:1.27cm;margin-right:0cm;"><span style="color:#ff9c00;">'''MEGASCII.sfp'''</span> for the head surface points, and</div> | <div style="margin-left:1.27cm;margin-right:0cm;"><span style="color:#ff9c00;">'''MEGASCII.sfp'''</span> for the head surface points, and</div> | ||
| − | <div style="margin-left:1.27cm;margin-right:0cm;"><span style="color:#ff9c00;">'''MEGASCII.pos'''</span> for the MEG sensors</div>* MEG coordinates will be correct. The sensor definition file specifies the sensors as planar gradiometers. | + | <div style="margin-left:1.27cm;margin-right:0cm;"><span style="color:#ff9c00;">'''MEGASCII.pos'''</span> for the MEG sensors</div> |
| + | * MEG coordinates will be correct. The sensor definition file specifies the sensors as planar gradiometers. | ||
* Where the auxiliary files came from will be recorded in the database. If you open the file again, the auxiliary files will be found automatically, without asking any questions. | * Where the auxiliary files came from will be recorded in the database. If you open the file again, the auxiliary files will be found automatically, without asking any questions. | ||
| − | |||
| Line 922: | Line 791: | ||
The files described in this example can be found in the ''Examples\Xtras\MEG+EEG'' subdirectory of the BESA Research installation folder. | The files described in this example can be found in the ''Examples\Xtras\MEG+EEG'' subdirectory of the BESA Research installation folder. | ||
| − | Here are two examples containing mixed MEG, EEG, and polygraphic channels:* Open a file using the ''File/Open'' command | + | Here are two examples containing mixed MEG, EEG, and polygraphic channels: |
| + | * Open a file using the ''File/Open'' command | ||
* Import a file using ''File/Import ASCII'' command | * Import a file using ''File/Import ASCII'' command | ||
| − | |||
| − | |||
| − | |||
| − | |||
In both cases | In both cases | ||
| − | |||
* The <span style="color:#ff9c00;">'''<nowiki>*.ela</nowiki>'''</span> file defines the channel labels. Based on the labels, BESA Research knows which channels are EEG and MEG. The remainder are classified as polygraphic channels. | * The <span style="color:#ff9c00;">'''<nowiki>*.ela</nowiki>'''</span> file defines the channel labels. Based on the labels, BESA Research knows which channels are EEG and MEG. The remainder are classified as polygraphic channels. | ||
* The <span style="color:#ff9c00;">'''<nowiki>*.pos</nowiki>'''</span> file defines the MEG sensor coordinates. The number of values on a line of this file (=9) defines the MEG as gradiometers. The relative locations of primary and secondary coils identify the gradiometers as planar. | * The <span style="color:#ff9c00;">'''<nowiki>*.pos</nowiki>'''</span> file defines the MEG sensor coordinates. The number of values on a line of this file (=9) defines the MEG as gradiometers. The relative locations of primary and secondary coils identify the gradiometers as planar. | ||
| − | |||
| − | |||
| − | |||
'''1. Example with ''File/Open''''' | '''1. Example with ''File/Open''''' | ||
| − | |||
| − | |||
* Open the file <span style="color:#ff9c00;">'''MEG+EEG.mul'''</span>. The ''Channel and digitized head surface point information dialog box'' will open automatically (unless the file has already been read once and the information is in the database). | * Open the file <span style="color:#ff9c00;">'''MEG+EEG.mul'''</span>. The ''Channel and digitized head surface point information dialog box'' will open automatically (unless the file has already been read once and the information is in the database). | ||
* You will see under ''‘internal data file information’'' that BESA Research finds 122 MEG sensors, and 162 channels in all. | * You will see under ''‘internal data file information’'' that BESA Research finds 122 MEG sensors, and 162 channels in all. | ||
| Line 948: | Line 808: | ||
* Under ‘''MEG sensors’'', the sensors have been identified as gradiometers. | * Under ‘''MEG sensors’'', the sensors have been identified as gradiometers. | ||
* The green tick at the top right of the window indicates that BESA Research classifies everything as OK. | * The green tick at the top right of the window indicates that BESA Research classifies everything as OK. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
'''2. Example with ''File/Import ASCII''''' | '''2. Example with ''File/Import ASCII''''' | ||
| − | |||
| − | |||
* Import the file <span style="color:#ff9c00;">'''MEG+EEGimport.asc'''</span>. Select <span style="color:#ff9c00;">'''MEG+EEGimport.fsg '''</span>as the target file (see ''“Example: ASCII Import”''). | * Import the file <span style="color:#ff9c00;">'''MEG+EEGimport.asc'''</span>. Select <span style="color:#ff9c00;">'''MEG+EEGimport.fsg '''</span>as the target file (see ''“Example: ASCII Import”''). | ||
* Select 320 Hz sampling rate, and 500 ms pre-stimulus time. Other selections in the dialog box should be ‘Multiplexed’, 1 bin/microvolt (this is interpreted as 1 bin/fT for MEG), 162 channels and 320 samples. | * Select 320 Hz sampling rate, and 500 ms pre-stimulus time. Other selections in the dialog box should be ‘Multiplexed’, 1 bin/microvolt (this is interpreted as 1 bin/fT for MEG), 162 channels and 320 samples. | ||
* The ''Channel and digitized head surface point information dialog box'' will open as above. | * The ''Channel and digitized head surface point information dialog box'' will open as above. | ||
Revision as of 09:31, 7 April 2017
Contents
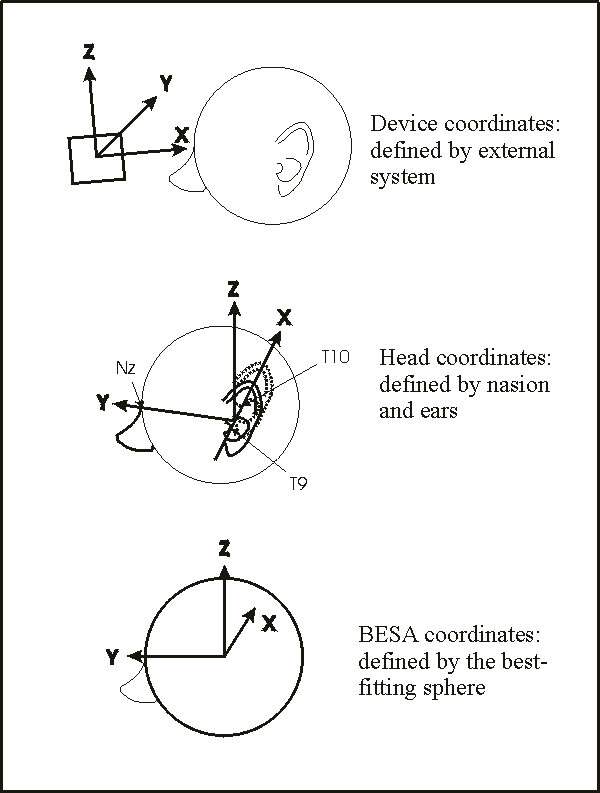
- 1 Working with Electrodes and Surface Locations
- 1.1 Introduction - Electrodes and Surface Locations
- 1.2 Working with auxiliary files
- 1.3 Coordinate systems
- 1.4 The Channel and Digitized Head Surface Point Information Dialog Box
- 1.5 General Reading Rules for Data Files and Auxiliary Files
- 1.6 Electrodes
- 1.7 3D Coordinates for Precise Analysis
- 1.8 Example: Defining Channel Labels
- 1.9 Example: Mixed EEG and Polygraphic Data
- 1.10 Example: EEG with Digitized Coordinates
- 1.11 Example: Polhemus Digitizer Data
- 1.12 Example: Digitization points with and without Fiducials
- 1.13 Example: ASCII Import
- 1.14 Example: MEG ASCII
- 1.15 Example: Reading combined EEG and MEG from an ASCII file
Working with Electrodes and Surface Locations
Introduction - Electrodes and Surface Locations
Here you will find out how BESA Research works with electrode and MEG sensor coordinates and labels, and how head surface points can be used to improve source modeling and coregistration of source models with the MRI. In most cases, electrode positions are sufficiently defined by their labels. The section "Electrode Conventions" lists the standard position which BESA Research assigns to EEG channels. In some cases, especially for larger electrode arrays or for MEG measurements, additional information is required to add sufficient information for mapping and for source analysis. The additional information is supplied in additional, auxiliary files which are read by BESA Research and associated with the data files. The auxiliary files, and how they are supplied to BESA Research, are described in this chapter.
Examples for using auxiliary files to define the 3D locations of electrodes are found in the chapter "Special Topics / Working with Electrodes... / Examples" in the online help.
Descriptions of file formats that BESA Research uses are given in the online help chapter "Special Topics / Working with additional files".
Working with auxiliary files
Data files come with varying amounts of prior information about electrode/sensor locations, depending on the recording system. BESA Research allows you to read auxiliary files that define additional information, such as channel labels, and coordinates of the electrodes, sensors, and other head surface points. The information is required for mapping and for source montages.
- Mapping. BESA Research uses spherical spline mapping. For this, electrode/sensor locations are projected onto a sphere. The minimum requirement is 10-10 or 10-20 labels: if only channel labels are available without additional information, BESA Research uses default spherical coordinates.
- Source modeling. Spherical coordinates of electrode locations are sufficient, but digitized locations are better. Digitized locations can be defined in the data file or in auxiliary files. BESA Research will use digitized head surface points (electrodes + additional points) to fit a sphere for the spherical model. Points anterior to the left and right preauricular points and below the plane formed by these points and the nasion are excluded when fitting the sphere.
Files can also be written, for instance for
- Source modelling with MRI coregistration. BESA Research allows for the export of surface points in a special format (sfh file) which can be read by the BESA MRI (or BrainVoyager) program. These are fitted to the head surface defined by BESA MRI (or BrainVoyager) in order to define rotation, translation, and deformation parameters required to coregister the coordinate systems (see "Integration with MRI and fMRi").
- Export of coordinates. Electrode, other surface point locations, and MEG sensor coordinates and other surface point locations can be written to ASCII files so that they can be reread when reading other files into BESA Research (e.g. ASCII files), or used by other programs.
Feedback and control over how these files are read is provided by
- the Channel and digitized head surface point information dialog box. This dialog is usually opened when you open a file for the first time. It allows you to specify the names of auxiliary files, and it makes initial checks on the files to see whether they are consistent with each other and with the data file. If the check is OK, you will see a green tick at the top right hand corner of the dialog box. If there are inconsistencies, the tick is replaced by a red exclamation mark. In this case, you will usually need to edit the auxiliary files or specify other files. The dialog box is not opened if the file is recognized to contain all the necessary information (files with the foc, fsg extensions), or if the program only finds a channel definition file (with extensions ela, elp, or elb). The dialog box is not opened if the data file has been opened before. You can always open the dialog box manually by specifying " File / Head Surface Points and Sensors/Load Coordinate Files.
- The log file (*_LoadFile.log). Coordinating the information between the data file and its auxiliary files can be a complex procedure. To help you check whether the coordination is being done properly, if you select the menu entry " Options / File / Generate Log " during File Open, BESA Research writes a log file with the same base name as the data file, appending "_LoadFile.log" to the base name, recording which files have been read, and some of the parameters that have been found. This file is created every time auxiliary files are read (e.g. on file open, when reading in channel configuration files, head surface point files, MEG sensor locations), or changed ("Edit / Channel Configuration").
- The log window. If there are inconsistencies during the processing of auxiliary files and 3D coordinates, a logging window is opened showing the information that would be written to the log file. You can read what has been done to help diagnose the problems. Select OK to continue in spite of the problems, or Reset to reject. Typing Reset also deletes the database files associated with the current data file, thus allowing you to start reading this file from scratch.
BESA Research remembers which auxiliary files are associated with the current file. When a data file is first opened, and BESA Research finds auxiliary files with the same base name as the data file, you will be asked if you want this file to be read. The decision you make will be recorded in the database for this data file. Next time the file is opened, the files will or will not be read, according to your previous decision. Similarly, when an auxiliary file is read using the menu, this is recorded in the database, and the file will be opened automatically next time the data file is opened. To override previous decisions, you must delete the database files (see the log window above) or change the entries in the Channel and digitized head surface point information dialog box (see the chapter "Electrode Conventions / Channel and digitized head surface point information dialog box").
Coordinate systems
We need to deal with four different coordinate systems. These differ in how the x, y, and z axes are defined, and in the units of measurement (e.g. mm, cm, m). The first three are illustrated in the following figure:
Device coordinates. These are the coordinates used by the recording system. The axes may be anywhere in relation to the head. For instance, in the Polhemus digitizer, the axes go through the magnetic field transmitter which is located somewhere outside the head. The units of measurement may be millimeters, centimeters, or meters.
Head coordinates. This coordinate system is defined by reference points on the head known as fiducials. The reference points are normally the nasion (Nz, NAS), the left preauricular point (T9, LPA), and the right preauricular point (T10, RPA). The x axis is defined by the line joining T9 and T10, positive towards T10. The y axis is defined by the line through Nz that is perpendicular to the x axis (positive towards Nz). The z axis is perpendicular to the x and y axes, and goes up out of the head in the vicinity of Cz. The units of measurement may be millimeters, centimeters, or meters. In BESA Research these are labelled with the prefix 'Fid', e.g. 'FidT9', 'FidNz'.
BESA Research coordinates. For dipole analyses the head model consists of a sphere. In the default situation where no digitized sensor information is available, the center of the sphere is defined by the crossing point between the lines joining T7 (=T3) and T8 (=T4) and Fpz and Oz.. The x axis is the T8-T7 line, positive at T8. The y axis is the Oz-Fpz line, positive at Fpz. The z-axis goes up out of the head through Cz. If digitized information is available, the axes are defined by the best fit between the idealized electrode locations and the real locations. The diameter of the sphere is also defined by the best fit. Units given in the display are in millimeters.
The center of the spherical model is on average about 4 cm above the origin of the Head Coordinates. If digitized surface points are available, the sphere is fitted to these points. Using a cot file, it is possible to override the fit and define your own head center. In conjunction with BrainVoyager, you can use the MRI to seed the location of the head center (e.g. a fixed distance anterior to the posterior commissure) and save it as a cot file. Using MRI coregistration (see "Integration with MRI and fMRi"), the center is placed between the anterior (AC) and posterior (PC) commissures, at the half-way point between the anterior and posterior points (AP and PP). Without coregistration, the center corresponds to a point 17.5 mm behind AC in the standard MRI head.
MRI coordinates. These are the coordinates used by BrainVoyager. These are defined by the MRI slices. Measurement units are millimeters.
The Channel and Digitized Head Surface Point Information Dialog Box
Many data formats read by BESA Research require additional information about data channel, which are specified by additional, auxiliary files. This dialog box allows you to specify which auxiliary files are read in to supplement the information in the data file.
The dialog box is opened automatically the first time a data file is opened, if
- auxiliary files other than a channel definition file (*.ela, *.elp, *.elb) are found
- no auxiliary files are found, and the data file was not written in compressed binary format (*.foc, *.fsg) by BESA Research
When a data file is closed, the information about which auxiliary files have been read is stored in the database. When the file is opened for a second time the dialog box is not opened automatically, because the information is assumed to be correct – the files are read automatically.
The dialog box can be opened manually by selecting "File / Head Surface Points and Sensors/Load Coordinate Files", or using the shortcut ctrl-L
The dialog box is divided into several sections:
- Internal data file information. Here you can see the file name, the originating system (file format), the name of the database file, if any, and the channel information as specified by the data file alone.
- Suggestions. This box makes suggestions about what needs to be filled in, e.g. "Please enter electrode thickness"
- Main feedback (top right hand corner). A green tick indicates that the currently selected data files are consistent among themselves and with the data file. A red exclamation mark indicates an inconsistency. Check the feedback texts in the subsequent sections for more information:
- Channel configuration file specification. If the channel labels and types defined in the data file ("Internal data file information") need to be changed, enter a file name here (*.ela, *.elp, *.elb). If a channel definition file exists with the same basename as the data file, or if a channel definition file has been specified previously (database entry exists), it will be selected automatically. To the right of the file name, feedback is provided about the number of channels and channel types found. If the labels are consistent with the data file, to the right the text "Good" is shown. If they are inconsistent, e.g. the file contains the wrong number of channel definitions, the text "Bad" is shown.
- Digitized head surface point specification. Here you may specify a file containing digitized electrode and other head surface points. Optionally, the information can be split into two files, containing the coordinates (*.sfp, .eps) and the coordinate names (.sfn). Alternatively, both labels and names can be contained in the coordinate file. If the files specify electrode coordinates, there must be a coordinate name for each electrode. The sequence may be different. BESA Research will use the names to assign each coordinate to the electrodes. Additional head surface points can have any other names. It is recommended that the first three digitized coordinates are the fiducials (fiduciary points), labelled "FidT9", "FidT10", "FidNz". If your electrode labels not follow the 10-10- or 10-20 standard (e.g. in high-density electrode recordings), it is recommended to tick the box "Electrode labels non -conforming to 10-10 standard". This will prevent BESA Research from using electrodes for an optimal rotation of the coordinate system which should not be used (e.g. A1, A2 which have known locations in 10-10, but are sometimes used in a nomenclature outside of 10-10). The example below shows the sphere adaption for an example data set with and without taking this into account. The right picture shows that when discarding the non-conforming electrodes, the fiducials are correctly placed along the x any y axes.
(In the special case of Neuromag files with electrode channels, the data file contains head surface points with the wrong labels. Here you may provide a label file (*.sfn) without a corresponding digitized coordinate file.)
- Coregistration file. Here you may specify a file containing the coordinates of the head center (*.cot) or an MRI Coregistration File (*.sfh). Head center redefinition is only necessary if you
- MEG sensor specification. If the file contains MEG channels, you may enter the name of a sensor coordinate file (*.pmg, *.pos). This field is grayed if there are no MEG channels.
- Artifact coefficients file. If the data are to be artifact corrected, your pre-prepared coefficient file may be defined here. See the chapter "Artifact Correction". Selecting the file here is equivalent to loading the file using the menu entry "Artifact / Load".
For each of the selected files, make sure the radio button "Yes" is selected on the left-hand side of the dialog box. If files have been selected automatically, and you do not wish them to be read, select the "No" radio button.
If some of the settings are incorrect or the text "Bad" is shown, you may edit the auxiliary files (the file is opened with the NotePad program) or browse for another file by pressing the Edit or Browse buttons.
After you have entered the required information, and the green tick at the top right indicates consistency, press OK to continue. Press Cancel to ignore the current settings. Press Clear DB to delete the database files. Press Clear Events to delete the tag files (the part of the database that records events). Both Clear buttons close the currently-opened data file.
General Reading Rules for Data Files and Auxiliary Files
Auxiliary files can complement the information in the data file. Here we specify what happens when a data file is opened:
1. If the data file has been read previously, the database entry specifies which auxiliary files should be read. The file and the specified auxiliary files are opened and the data are displayed.
2. If a) there is a channel definition file (*.el?) with the same basename as the data file, and
b) this file includes spherical coordinates for the EEG channels (including labels with entries in the default.ecd file), and
c) there are no other auxiliary files with the same base name, the file will be opened and the data displayed. If files with the same basename are not found, BESA Research will look for files with the basename “default” (e.g. default.ela) in the data folder. If such files are not found, BESA Research will look for files with the basename “default” one folder above (e.g. ..\default.ela).
3. If the data file has been written in binary format (*.foc, *.fsg) by BESA Research (after Jan.2000), the file will be read, and all information is assumed to be complete. The file is opened and the data are displayed.
4. In all other cases, the Channel and digitized head surface point information dialog box will be opened for you to specify and check auxiliary files. Auxiliary files with the same base name as the data file will be specified in the text boxes for file names. If files with the same basename are not found, BESA Research will look for files with the basename “default” (e.g. default.ela) in the data directory. If such files are not found, BESA Research will look for files with the basename “default” one directory above (e.g. ..\default.ela). Otherwise the text boxes will be left blank.
5. Auxiliary files can be specified at a later time by selecting File/Head Surface Points and Sensors/Load Coordinate Files. The Channel and digitized head surface point information dialog box will be opened.
Electrodes
Electrode Conventions
BESA Research adheres to the 10/20 and to the new 10/10 standard of the IEF (international EEG Federation). BESA Research will recognize the labels defined by these standards. The labels are stored in most EEG file headers. Otherwise, or in the case of erroneous labeling or sequencing of the recording channels, you may edit the channel labels and/or coordinates, or you may read an electrode file stored previously on disk. In addition to the 10/20 and 10/10 standard labels BESA Research recognizes the following labels: M1, M2 (left, right mastoids), SP1, SP2 (sphenoidal), CB1, CB2 (cerebellar), Chin, Neck, LO1, LO2 (lateral ocular), SO1, SO2 (supra-ocular), IO1, IO2 (infra-ocular). BESA Research will translate all the labels into spherical coordinates for spherical spline interpolation, mapping and source imaging. The following assignments are stored in the file default.ecd in the BESA folder:
Electrode labels in the file default.ecd and their spherical coordinates. 10-20 electrodes are shown in red and italic.
The spherical coordinates are defined in degrees by the azimuth (from Cz, positive = right, negative = left hemisphere) and the latitude (counterclockwise from T7/T3 for left and from T8/T4 for right hemisphere) of each electrode. Please do not modify the existing labels or coordinates in this file, because this would adversely affect the interpolated (virtual) montages, the maps and the source montages and source images in BESA Research. However, you may add additional labels for scalp electrodes at the end of this file if needed (up to a total of 196). When you edit the electrode configuration or read in electrode files, BESA Research may replace the 10/20 standard labels T3, T4, T5, T6 by their new 10/10 equivalents T7,
T8, P7, P8. However, in the initialization file BESA.ini you can reset to the old 10/20 standard by relabeling T7=T3, P7=T5, T8=T4, P8=T6 under the heading [Electrodes]. You may use the same feature to assign appropriate labels to the X1..X8 channels which exist in many systems, e.g. X1=EKG1 etc.
Recommendations for electrode placement
For source montages and source analysis two principles are important:
- Covering of the lower head with inferior electrodes to record activity from the inferior surfaces of the brain, especially from the basal temporal lobe, from the temporal pole, from orbito-frontal cortex, and from basal occipital and cerebellar areas.
- Equal spacing of the electrodes over the whole head to cover all brain areas.
In the following montage EEGxx the number xx indicates the number of electrodes.
EEG25 - Minimum 10-20 configuration including inferior electrodes
This covers the 19 standard 10-20 electrodes:
Fp1, Fp2, F7, F3, Fz, F4, F8 ....
plus 6 inferior electrodes on both sides:
F11, A1, P11, F12, A2, P12
with a recommended continuation of the 20% distances, i.e. use F11 instead of F9, P11 instead of P9, A1 instead of T9 to have a wider coverage of the inferior head. A1 / A2 may be replaced by T9 / T10 (or FT9 / FT10) for convenience and comfort.
Left: recommended configuration for 25 electrodes. Right: left temporal basal activity mapped with 25 electrodes.
EEG33 - Additional 10-10 electrodes within the major squares
To the above electrodes add the following 8 intermediate electrodes:
FC5, FC1, FC2, FC6, CP5, CP1, CP2, CP6
Left: recommended configuration for 33 electrodes. Right: left temporal basal activity mapped with 33 electrodes.
EEG35 - Additional supraorbital electrodes for better EOG separation
SO1, SO2
EEG37 - Wider inferior coverage at interlaced 20% distances
Continue 20% down from F7, FC5, CP5, P7 etc. and use the following 8 inferior electrodes instead of 6:
F11, FT9, TP9, P11, F12, FT10, TP10, P12
EEG41 – Improved frontal and occipital coverage
Additional electrodes halfway between Fz and Fp1 / FP2 and Pz and O1 / O2:
AF1, AF2, PO1, PO2
EEG43 – Inferior chain with 5 electrodes including A1 / A2
F11, FT9, A1, TP9, P11, F12, FT10, A2, TP10, P12
EEG43 represents the widest coverage with relatively even spacing.
Left: recommended configuration for 43 electrodes. Right: left temporal polar activity mapped with 43 electrodes.
EEG64-256
With 64 or more channel caps, it is similarly recommended to use a sufficient number of inferior electrodes all around the head. At least 4 inferior temporal electrodes on each side and additional electrodes above or below the eyes (outside of the cap) are suggested.
Editing the channel configuration
Only use the channel configuration editing facility if the electrodes or the common reference have not been correctly defined by your digital EEG system, or if you want to define specific spherical coordinates for your scalp electrodes. It is your responsibility to check and provide the correct sequence of electrode labels in correspondence with the sequence of channels in the EEG data file. If these sequences do not match exactly, errors will occur in the computation of maps, source images and interpolated montages.
We will use the example EEG file eeg2.eeg in the subdirectory Examples/EEG-Focus of the BESA Research directory to explain the editing of electrode labels and coordinates:
1. Select File, then click on Open, or click on eeg2.eeg if this file is contained in the list of currently selected EEG files. 2. Select Edit, then click Channel Configuration. The dialog box shown below will appear.
At the upper left of the figure you see the dropdown menu after selecting the File menu in the dialog box. This menu allows to edit a new (New) or an existing (Open) electrode file and to save the changes to the same (Save) or a different (Save As) file. Normally, it will not be necessary to use this menu. The control fields on the right will be sufficient. If you type 'Ok', you will be given the option of saving the changes to a file.
3. Click on Reload org. Labels to reread the original labels as stored in the file header of eeg2.eeg. BESA Research quits editing and redisplays the EEG. Repeat step 2 and select "Edit / Channel Configuration" again. Note: The button Reload org. Labels is not available if there are no labels in the file header. 4. Click on the empty space of the scroll bar below the scroll button and on the down arrow of the scroll bar to display the remaining electrodes in the file. 5. Click on electrode R (line 32), then on the button Delete Electrode to remove the associated channel, which does not contain any signal. Note that you may not omit intermediate channels, even if they do not exhibit signals, because the correct correspondence between the series of electrodes and the EEG channels will not be maintained. Use the "Edit / Bad Channels" menu to disable artifactual or empty channels. 6. Click on EOG (line 30) and change the entry to EOG1. Do not type <Enter>, but click on the next or a different electrode box to accept the changes. 7. Double click on EOG (line 31). This will highlight the entry. Simply type the new name EOG2, and note that the old label is replaced when highlighted. Electrodes 30 and 31 are now defined as distinct electrodes. Next, we want to replace the label T10 by A2. 8. Click on the label T10 (line 24). Then click on the drop down arrow right of the highlighted label to obtain the list of default scalp electrodes (read from default.ecd and sorted alphabetically). Type A to jump to the electrodes beginning with letter A (see below). Type 2 or click on A2 in the list. Click on the type box (Scalp channel) to close the list and display the new entry in line 24. Note that this is the most convenient way to edit an electrode label.
9. Exercise: repeat step 8 to replace T9 by A1. Restore labels T10 and T9 in lines 24 and 21.
10. Replace SO1 and SO2 (supra-orbital) by PSO1 and PSO2 and note that these electrodes are changed to the 'Polygraphy' type, because no coordinates are associated with these labels.
11. After you click 'OK', the box Write Channel Configuration File will appear and display a name for the current electrode file. By default, the BESA electrode file path and current file name will be used and supplemented by the extension '.elb'. The electrode file path may be set in the BESA.ini file [Defaults] section under ElectrodeFilePath. If no electrode file path is specified in the BESA.ini file, the default electrode file path Montages\Channels is used. Simply click 'OK' or type <Enter> to save the changes to this file, or select a new file name and/or path, if you do not want to store the electrode file in the BESA Research electrode file directory.
Note that by using the default 10/10 labels (see chapter "Electrode conventions") you specify that the associated electrode is a scalp electrode. Hence, different labels must be used for polygraphic, intracranial or MEG channels. After you have entered a new non-scalp label, you may select the type of the electrode/channel amongst the different groups (Polygraphy, Intracranial, MEG Channel) from the drop down list in the 'Type' box. This will allow for using separate selection and scaling facilities of the channel group control push-buttons at the right of the screen (All, Scp, Pgr, Icr, MEG). If you have entered a new non-scalp label and select the type Scalp Channel, or if you click on the 'Advanced>>' field, boxes will appear to enter the spherical coordinates (azimuth and latitude) of this electrode (cf. Fig. 6.5). These features may be used to specify non-standard scalp electrodes. Please check the earlier sections of this chapter for electrode conventions. You may view the locations of the scalp electrodes on the head schemes in the mapping window. Select Show Electrodes in Maps in the "View / Options" menu.
Hint: If you want to specify the spherical coordinates of an electrode which is close to a standard electrode, click on the 'Advanced >>' field, enter the label of the standard electrode and append a single quotation mark. This will specify that the electrode is close to the labeled location but has different coordinates. The Scalp Channel type will not be replaced by Polygraphy. Then edit Azimuth and Latitude. This convention is used by BESA Research when reading electrode coordinate files (*.elp), e.g. from the BESA program. The coordinates are read and compared with the default coordinates to assign the closest label. Then a single quotation mark is appended to the label, and the coordinates are assigned as specified in the .elp file. For example, open segm1.eeg in the Examples\EEG-Focus subdirectory.
Note that the file segm1.elp is searched for automatically in the directory of the data file when opening the data file.
Edit Common Scalp Reference
There is a separate line at the bottom in the Channel Configuration dialog box to enter the label and coordinates of the Common Scalp Reference electrode. If this is specified and enabled (click on field Enabled), the information provided by the fact that all scalp electrodes were recorded against a common recording reference will be used for mapping, source imaging and virtual montages. This information will be lost if the common reference has not been specified or if a combination of electrodes has been used as reference during recording. Specify the Common Scalp Reference electrode only if all electrodes have been referenced to the same single electrode and if a standard 10/10 location has been used for the common recording reference.
Note that BESA Research cannot process digital EEG data correctly if there is no common recording reference, and if different recording references were used for the various scalp electrodes. For intracranial and polygraphic channels different references may be used. It is preferable to use the common reference also for electrode channels near the eyes, because these electrodes provide valuable information for mapping, source imaging and interpolated montages. The traditional bipolar channels (e.g. horizontal and vertical EOG) may be reconstructed digitally using the Selected Channels group or user-defined montages.
3D Coordinates for Precise Analysis
Introduction - Working with Digitized 3D Coordinates
Working with digitized electrode coordinates usually requires reading in additional (auxiliary) files. The procedure is described in the chapter "Working with auxiliary files".
Data reading rules for EEG
This section explains which additional files are read, or which files have to be read in order to provide the necessary information for mapping and source montages.
Assume file name is datafile.xxx. datafile is the base name, .xxx is the extension. Replace the text datafile by the base name of your own file, and the extension xxx by the extension of your own file.
Channel definitions for EEG:
Labels have 10-10 names: default locations will be used.
Labels do not have 10-10 names: Channels are interpreted as polygraphic. Mapping is not possible without one or more of the following additional files.
Define channel names and types. File datafile.ela, datafile.elp, or datafile.elb exist, or files default.ela, default.elp, or default.elb exist, or files ..\default.ela, ..\default.elp, or ..\default.elb exist (i.e. files with basename default one folder above the data file): Channel names and types will be replaced by those defined in this file, in order of occurrence. The ela file contains just labels and, optionally, types. The elp file contains spherical coordinates and can contain labels and types. The elb file contains the same information in binary format. See chapter "Working with additional files / Channel Definition File Conventions".
Define order in which electrodes were digitized. File datafile.sfn exists, or file default.sfn exists, or file ..\default.sfn exists (i.e. file default.sfn one folder above the data file): electrode names are supplied in the order in which coordinates were supplied in the sfp file. These names must match with the names supplied in the data file or defined in the ela/elp/elb file. BESA Research uses this to sort coordinates into the order of channels in the file. If fiducials exist, they should be defined on the first three lines. If they do not exist, BESA Research will simulate them (so that it can define the head coordinate system). See chapter "Working with additional files / sfn (surface point name) file".
Define electrode coordinates. File datafile.sfp exists, or file default.sfp exists, or file ..\default.sfp exists (i.e. file default.sfp one folder above the data file): electrode coordinates will be replaced/defined by the coordinates defined in this file. If datafile.sfn does not exist, labels can also be defined in this file. If fiducials exist, they should be defined on the first three lines. If they do not exist, BESA Research will simulate them. See chapter "Working with additional files / sfp (surface point coordinate) file".
Define coregistration information. File datafile.sfh exists, or file default.sfh exists, or file ..\default.sfh exists (i.e. file default.sfh one folder above the data file): head center and relative position of the unit sphere with respect to the head coordinate system is determined by the coregistration between EEG and MRI. See online help chapter "Integration with MRI and fMRI".
Define head center. No coregistration file exists (*.sfh, see above). File datafile.cot exists, or file default.cot exists, or file ..\default.cot exists (i.e. file default.cot one folder above the data file): head center as computed by fitting a sphere to the surface points is replaced by the head center coordinates contained in this file. See chapter "Working with additional files / cot (Head center) file".
Data reading rules for MEG
Assume file name is 'datafile.xxx'. datafile is the base name. xxx is the extension. Replace the text datafile by the base name of your own file, and the extension xxx by the extension of your own file.
Here we consider cases a) MEG alone, b) MEG+EEG.
Automatic procedure:
Labels have names defined in the files bti.ecd or nmag.ecd. Channels are interpreted as MEG. However, sensor locations and head surface point locations must be defined in additional files as described below. Mapping and source analysis are not possible without one or more of the following additional files.
Define channel names and types. File datafile.ela, datafile.elp, or datafile.elb exist, or files default.ela, default.elp, or default.elb exist, or files ..\default.ela, ..\default.elp, or ..\default.elb exist (i.e. files with basename default one folder above the data file): Channel names and types will be replaced by those defined in this file, in order of occurrence. The ela file contains just labels and (optionally) channel types. The elp file contains spherical coordinates and can contain labels and types. The elb file contains the equivalent information in binary format. See chapter “Electrode file conventions and formats.”
MEG+EEG. Define order in which electrodes were digitized. File datafile.sfn exists, or file default.sfn exists, or file ..\default.sfn exists (i.e. file default.sfn one folder above the data file): electrode names are supplied in the order in which coordinates were supplied in the sfp file (or in the location descriptor in the data file: e.g. Neuromag). These names must match with the names supplied in the data file or defined in the ela/elp/elb file. BESA Research uses this to sort coordinates into the order of channels in the file. See chapter “Working with additional files/ sfn (surface point name) file”.
MEG+EEG. Define head surface point/electrode coordinates. File datafile.sfp exists, or file default.sfp exists, or file ..\default.sfp exists (i.e. file default.sfp one folder above the data file): electrode coordinates will be replaced/defined by the coordinates defined in this file. If datafile.sfn does not exist, labels can also be defined in this file. See chapter “Working with additional files/ sfp (surface point coordinate) file”. The labels of electrode coordinates must match to those defined for the data channels. BESA Research will use the labels to associate coordinates with the correct channel.
Define sensor coordinates. File datafile.pos or datafile.pmg exists, or file default.pos(.pmg) exists, or file ..\default.pos(.pmg) exists (i.e. file default.pos(.pmg) one folder above the data file): coordinates are defined in this file. The convention is that pos files contain gradiometer coordinates and pmg files contain magnetometer coordinates. This is not necessary for the program to read in values properly: the program makes its decision about the sensor type on the basis of the number of coordinate values on one line in the file (6 = magnetometers, 9 = gradiometers). See chapter “Working with additional files/ pos or pmg (MEG sensor coordinate) file”.
Define coregistration information. File datafile.sfh exists, or file default.sfh exists, or file ..\default.sfh exists (i.e. file default.sfh one folder above the data file): head center and relative position of the unit sphere with respect to the head coordinate system is determined by the coregistration of the head coordinates with MRI. See (online) help chapter "Integration with MRI and fMRI".
Define head center. No coregistration file exists (*.sfh, see above). File datafile.cot exists, or file default.cot exists, or file ..\default.cot exists (i.e. file default.cot one folder above the data file): head center as computed by fitting a sphere to the surface points is replaced by the head center coordinates contained in this file. See chapter "Working with additional files / cot (Head center) file".
Reading MEG files in ASCII format
BESA Research uses labeling or channel type definitions to decide whether channels are EEG or MEG. Based on the labels defined for the channels, or the type specified by the channel definition file, the program will try to find auxiliary files that define electrode coordinates or MEG sensors.
BESA Research uses four files to make its decision:
- .ela/.elp The channel type defined here overrides definitions in *.ecd (below).
- default.ecd defines electrode labels and default spherical coordinates based on the 10-20 and 10-10 naming system
- bti.ecd defines labels and default spherical coordinates for the BTi whole-head system
- nmag.ecd defines labels and default spherical coordinates for the Neuromag whole-head system
If the program finds a label in default.ecd, the channel will automatically be defined as EEG. If not, if it finds a label in one of the other files, the channel will be defined as MEG. If it doesn't find the label anywhere, the channel will be defined as Polygraphic.
The spherical coordinates defined in these files are sufficient for mapping the data. Auxiliary files defining the real sensor coordinates are required for source analysis.
Defining default label and coordinate file for a new MEG system
When preparing an MEG from a system other than BTi-WHS or Neuromag for import to BESA Research, you should edit either bti.ecd or nmag.ecd to conform with your system. If sensor files (*.pos/*.pmg) are always available for your files, the coordinates in the ecd files are irrelevant: all you need do is define the labels for your own MEG system or use the labels as already defined.
Files to prepare for reading in each data file
Each auxiliary file should have the same base name as your data file.
Define channel labels. There are several possibilities:
- Generate your data file according to the BESA avr format or the ASCII multiplexed format. Labels are listed in the second line of the file.
- Generate a label file (extension .ela) with one MEG channel label per line (matching with your ecd file as defined above) or with the type "MEG" and a label for each line.
Label definitions are also possible using elp or elb files, but the above two solutions are recommended because they are the simplest.
Define sensor coordinates. Generate a pos or pmg file. Make sure that the number of sensors matches with the number of MEG channel definitions in your data file.
Define fiducials and other head surface points. Generate an sfp file. The first three lines define the fiducials. Subsequent lines define additional surface points.
Define coordinates of the center of the head. Generate a cot file. If this file is absent, BESA Research generates the coordinates by fitting a sphere to the head surface points.
Note that all coordinates should be within the same frame of reference, i.e. the same coordinate system. Units must be in meters, centimeters, or millimeters.
Example: Defining Channel Labels
The files described in these examples can be found in the .\Examples\Xtras\EEG+Channel Labels subdirectory.
The simplest way to define electrode coordinates is to use BESA Research’s default settings (defined in the file, default.ecd). In this case, you only need to provide a list of channel labels. If a channel label is defined in default.ecd (i.e. if the labels belong to the 10-20 or 10-10 system), BESA Research will recognize the channel as EEG, and will allocate 3D coordinates.
Labels are not always supplied correctly in the data file. You can override the internal labels in several ways:
- Read the data file, and then use "Edit / Channel Configuration" to redefine the channels. The configuration is stored in a file with the name basename.elb (for binary data) or basename.elp (for ASCII data), where basename is the base name (name without the extension) of your data file.
- Prepare a label file (with the extension ela) containing a list of labels. This can also specify channel types (e.g. EEG, Polygraphic, Intracranial, MEG).
- Prepare a file (with the extension elp) containing spherical coordinates of the channels. This is the method used with the previous version of BESA Research. If the file doesn’t contain labels, labels are allocated based on their proximity to the 10-20 or 10-10 definitions in default.ecd.
The following examples illustrate the above three methods. The input files are all in BESA avr format, although these examples apply to all EEG data formats in which only EEG channels exist.
If the data file contains polygraphic or other types of non-EEG channel, the types need to be defined. See the EEG+Polygraphic channels example. MEG is a special case, because the sensor coordinates need to be defined. See the MEG ASCII and the MEG+EEG examples.
Example 1. EEG file containing wrong labels – use Edit/Channel Configuration to redefine labels
- The avr file, EEGwithLabels.avr, contains the EEG labels. Channels 4 and 14 have been mislabeled – the labels need to be swapped.
- Open the file with File/Open (Select file type BESA avr. Find the correct directory Xtras\EEG+Channel Labels).
- The file should open correctly, displaying 32 channels of EEG.
- The channel coordinates can be viewed by typing the ‘V’ key (make sure the cursor is off). There will be a 3D display of the electrodes. Clicking on an electrode will display the label and the coordinates.
- In this file, channels 4 and 14 have been mislabeled as P3 and F3. In fact, the labels should be the other way around. We will now correct this:
- Select Edit / Channel Configuration.
- Type ‘F3’ into the label for channel 4, and ‘P3’ into the label for channel 14. Then type ‘OK’. The new channel configuration will be saved in EEGwithLabels.elp.
- In the data display, the labels of channels 4 and 14 will now be displayed correctly.
- Close the file (File/Close) and open it again. Note that the labels are still correct. This is because the new channel configuration file, EEGwithLabels.elp, is read automatically.
Example 2. EEG file with no labels – channel labels in auxiliary file
- The avr file, EEGwithoutLabels.avr, has no labels.
- EEG labels are defined in EEGwithoutLabels.ela. This is read automatically when the file is opened.
- In this example, labels are correct. Each label in the ela file is on one line:
Example 3. EEG file with no labels – channel labels derived from spherical coordinates
- The avr file, EEGwithSphericalCoords.avr, has no labels.
- Spherical coordinates are defined in the elp file, EEGwithSphericalCoords.elp. This contains spherical coordinates (theta and phi) and no labels:
- When the data file is opened, the elp file is read automatically, and BESA Research uses the tables in default.ecd to assign channel labels. To indicate that it has assigned user defined coordinates and matched with the closest standard electrode, BESA appends an apostrophe (‘) to each label:
- We advise to assign specific labels as well as spherical coordinates if you want to use your own spherical coordinate system, e.g.:
Example 4. EEG file with no labels – channel labels not in basename.el?
- The avr file, EEGnoLabelsNoElaFile.avr, has no corresponding ela, elp, or elb file, i.e. no file with the same base name and the el? extension.
- When you open the file, BESA Research will ask for a channel configuration file. The File Open dialog will select the directory .\Montages\Channels. The idea is that standard (= frequently used) electrode configurations should be kept in this directory.
- Select the file XtrasExample.ela.
- Close the data file and reopen it. The file will open with the correct labels. In the BESA window title you will see that the file EEGnoLabelsNoElaFile.elp has been read automatically. This file was created when XtrasExample.ela was read.
Example: Mixed EEG and Polygraphic Data
The files described in this example can be found in the .\Examples\Xtras\ EEG+Polygraphic subdirectory.
The data are in EEG+Polygraphic.avr.
The third channel is defined as polygraphic in the EEG+Polygraphic.ela file:
The prefix "POLY" specifies that the channel is polygraphic. Most other channels are interpreted as EEG because the labels are known in the 10-20 system.
Similarly, channel 31 is defined as intercranial, using the prefix "ICR".
Note that you can also define channels as EEG by specifying the "EEG" prefix (e.g. "EEG E1". This is useful if there are many more channels than are defined in the 10-10 or 10-20 systems, and if the channel coordinates are defined.
Example: EEG with Digitized Coordinates
The files described in this example can be found in the .\Examples\Xtras\ EEG+Digitization Points subdirectory.
In the previous examples, we have illustrated how to assign labels to channels using channel definition files. In those examples, only spherical coordinates were defined. Here we will show how to read digitized surface points into BESA Research, using the surface point (sfp) coordinate file and the surface point name (sfn) file.
The principles of defining digitization coordinate files are:
- The labels in the sfp/sfn file combination are used to assign coordinates to electrodes. Thus, if a coordinate has the name ‘Fz’ it will be assigned to the channel with the label ‘Fz’.
- In consequence, digitization of surface points can be in a different order to the sequence of channels in the data file. Matching to channels is done by comparing the labels.
- We recommend that the fiducial points, nasion, left preauricular point, right preauricular point be digitized. If you do not digitize them, BESA Research will simulate these locations (see “Example: Digitization points with and without Fiducials”). Fiducial points, labeled FidNz, FidT9, FidT10 should be the first three coordinates in the sfp file.
- As with the channel definition files, it is easiest for BESA Research if you name the sfp/sfn files using the base name of the data file, e.g. if the data file is named doodah.avr, name the sfp file doodah.sfp and the sfn file doodah.sfn.
- You specify the files to be read in the Channel and digitized head surface point information dialog box.
See “Example: Polhemus Digitizer Data” for a discussion of how to format the files originating from Polhemus and other digitizers.
Example 1. EEG file containing labels, sfp file containing coordinates, sfn file containing coordinate names
- The avr file, EEGdigitized1.avr, contains the channel labels. Therefore, we don’t need a channel definition file.
- The sfp file, EEGdigitized1.avr, contains digitized coordinates of electrodes and of additional surface points. The labels in the file do not correspond to the electrode labels in the avr file.
- The sfn file contains the corrected labels (1 line for each corresponding line in the sfp file). Now it is possible to match up electrode labels with the labels in the avr file.
- Open the data file, EEGdigitized1.avr. The Channel and digitized head surface point information dialog box will open automatically.
- Note the green tick mark at the top right of the dialog box. This is feedback to say that coordinates of all 32 electrodes have been found.
- Look at the entry ‘Digitized head surface points’. Here you will see that the sfp and the sfn files have been read automatically (because of the common base name). There are 51 locations. Note that the digitizer file can contain many more locations than the electrodes. BESA Research uses the locations for fitting the sphere of the spherical head model in source analysis. BESA Research can export these locations for coregistration with the MRI.
- Define the electrode thickness as 6 mm (at the right of the ‘Digitized head surface points’ box. This is the distance of the digitized point on the electrode to the surface of the head.
- Press ‘OK’ in the dialog box and view the coordinates by pressing the ‘V’ key.
Example 2. EEG file without labels, channel labels in ela file, surface point coordinates and names in sfp file
- The avr file, EEGdigitized2.avr, has no channel label. Therefore, a label file is required. Here, the label file is EEGdigitized2.ela.
- The sfp file, EEGdigitized2.sfp, contains digitized coordinates of electrodes and of additional surface points. The labels are defined correctly in the sfp file, i.e. for every EEG channel label there is a corresponding coordinate. Therefore, no sfn file is required.
- When you open the file, don’t forget to define the electrode thickness as 6 mm in the Channel and digitized head surface point information dialog box.
Example: Polhemus Digitizer Data
The files described in this example can be found in the .\Examples\Xtras\ EEG+Digitization Points subdirectory.
Data from the Polhemus (other digitizers too) may often not fit the format BESA Research requires for the surface point file. Note that Polhemus data can be exported directly into BESA-compatible sfp-files using the LOCATOR software.
BESA Research requires either
- just the cartesian coordinates (x, y, z) values -- one set of coordinates per line. In this case, labels must be defined in a parallel surface point name file, e.g.
- the cartesian coordinates plus a label. The label can be in front of or behind the coordinates on the line, e.g.
Here is an example of a few lines of a (sfp) file that are not read correctly by BESA Research:
What is wrong?
- First, some of the points are just numbered. These numbers don't tell BESA Research which electrode channel to which the coordinates should be assigned – assignments should be via channel labels and not numbers.
- Second, Nz, T9, T10 define the fiducials. Instead, the labels FidNz, FidT9, FidT10 are required (prefix "Fid").
What should be done? Probably the best way is
a) keep only the coordinates in the *.sfp file:
b) prepare a surface point name file (*.sfn) containing the corresponding labels:
Keeping labels and coordinates separate means that the label file needs generating only once. The coordinate file is different for each subject.
Alternatively, if your digitizer program attaches the labels correctly to the coordinates, then you can prepare the sfp file like this:
Example: Digitization points with and without Fiducials
We recommend that if electrodes are digitized, you should also digitize the three fiduciary points: Nasion, and left and right preauricular points. We refer to these points as "fiducials". We name them "FidNz", "FidT9", and "FidT10".
If you do not digitize these points, BESA Research will simulate them, i.e. it will generate the points where it expects them to be, based on the fit of a sphere to the existing points, and on the names of surface points of known locations. "Known locations" means: the surface point name must be a 10-20 or 10-10 electrode name (e.g. "Cz" -- arbitrary labels, such as "E10" is not a known location). Therefore, BESA Research requires that at least 3 surface points with known labels are defined.
In a file containing digitization points (*.sfp), the fiducials should be the first three sets of coordinates, i.e. the first three lines of the file. The remaining coordinates in the file can be electrode (or other surface point) coordinates, in any order. The assignment of electrode coordinates to data channels is achieved by matching the coordinate labels to data channel labels.
Consequences of omitting fiducials
When these files have been read into BESA Research, look at the head surface points in 3D using File/Head Surface Points and Sensors/View (or use the shortcut ‘V’). You will see small differences in fiducial locations between the real and the simulated locations. You can expect very slight influences on the results of source modeling (the spherical head may be rotated slightly, although the head center and radius will be identical), and output of source locations in head coordinates will be different, because these coordinates are based on fiducial locations (see chapter “Working with Electrodes and Surface Locations/ Coordinate systems”).
Example: ASCII Import
The files described in this example can be found in the .\Examples\Xtras\ASCII Import subdirectory.
When should the Import ASCII function be used? If you have data in BESA Research average referenced or multiplexed format, use the Open File function to read in a file directly. If you have data in a different ASCII format, BESA Research offers a flexible import function to import data from an array of numbers in an ASCII file.
The array can be vectorized (one channel, all time points, per line) or multiplexed (one time point, all channels, per line). These alternatives are illustrated in the two example files vectorized.asc and multiplexed.asc, and in the tables below:
Vectorized array:
| channel 1, time 1 | channel 1, time 2 | channel 1, time 3 | channel 1, time 4 |
| channel 2, time 1 | channel 2, time 2 | channel 2, time 3 | channel 2, time 4 |
| channel 3, time 1 | channel 3, time 2 | channel 3, time 3 | channel 3, time 4 |
| channel 4, time 1 | channel 4, time 2 | channel 4, time 3 | channel 4, time 4 |
Multiplexed array:
| channel 1, time 1 | channel 2, time 1 | channel 3, time 1 | channel 4, time 1 |
| channel 1, time 2 | channel 2, time 2 | channel 3, time 2 | channel 4, time 2 |
| channel 1, time 3 | channel 2, time 3 | channel 3, time 3 | channel 4, time 3 |
| channel 1, time 4 | channel 2, time 4 | channel 3, time 4 | channel 4, time 4 |
BESA Research needs channel labels. If the labels are in the 10-20 or 10-10 system, BESA Research will assign the channels default coordinates. This is the minimum requirement to be able to map EEG.
If you have 3D digitized coordinates, these can also be specified in ASCII files. This is described under the chapter “Example: EEG with Digitized Coordinates”.
Example 1. Vectorized data
- The file vectorized.asc contains the data. The file should be imported via File/Import ASCII File.
- First you will be asked for a name for the binary target file. The name vectorized.fsg is suggested. You may accept this name by pressing ‘OK’ or choose an alternative name. Note that if the file already exists, the imported data will be appended to the file.
- Next, the ASCII File Properties dialog box will open. First select ‘Vectorized’, and make sure the subsequent entries are correct:
- Next, the Channel and digitized head surface point information dialog box will open. In the ‘Channel configuration’ box, the label file, vectorized.ela, will be detected automatically. Automatic detection occurs when the label file has the same base name as the data file (in this case, vectorized). To the right of the file name is a summary of channel types: 32 channels found, 30 are EEG, 1 is intercranial, 1 is polygraphic.
Note the green tick at the top left of the dialog box. This indicates that BESA Research thinks that it has sufficient information to read the file, and map and do source analysis on the data.
- To see how channel types are specified in vectorized.ela, click on the Edit button to view the file with the Notepad program. Here you will see that most channels have 10-20 electrode names. Channel 3 has the prefix ‘POLY’, specifying that this channel is polygraphic. Channel 31 has the prefix ‘ICR’, specifying that this channel is intercranial. Close Notepad, and click ‘OK’ in the dialog box.
- A final dialog box asks for a Segment Comment. This is a label that will be displayed in the resulting file. The label is particularly useful if you import several ASCII files into one target file. Each segment is then easily identified by its own label.
Example 2. Multiplexed data
- This example is similar to Example 1. In this case, import the file multiplexed.asc, and select ‘Multiplexed’ in the ASCII File Properties dialog box. Other settings in the dialog box stay as they were.
Notes
- The numbers in the source files can be split into several lines per channel or per time point. Then you will have to enter the correct number of time points and channels in the dialog box. In the present examples, the lines are not split (the vectorized file has all 640 time points in each line, and the multiplexed file has all 32 channels in each line). In this case, BESA Research selects the correct numbers of time points and channels automatically.
- If you have digitized coordinates, these can be specified in the Channel and digitized head surface point information dialog box. Since the procedure is the same as when reading data, this is described elsewhere under “Example: EEG with Digitized Coordinates”.
Example: MEG ASCII
The files described in this example can be found in the \Examples\Xtras\ MEG ASCII subdirectory of the BESA Research installation folder.
Multiplexed MEG ASCII file with labels in the header (med.mul)
For reading MEG data, BESA Research expects
- Correct channel definitions, i.e. channels should be defined as MEG.
- Head surface points.
- Sensor coordinates, in the same coordinate system as the head surface points.
- Optionally, you can define the coordinates of the center of the head. This will be important if too few head surface points are available to specify where to place the spherical head used by BESA Research for source modeling, or if you want to use some external definition, e.g. from the MRI.
As with digitized EEG coordinates, we use the Channel and digitized head surface point information dialog box to specify the files which need to be read.
Example 1. File Open
- The file, MEGASCII.mul, contains MEG data in the ASCII multiplexed format. This format contains channel labels. The labels used are recognized by BESA Research as originating from the Neuromag system. They are therefore identified as MEG and do not need further identification.
- The sfp file, MEGASCII.sfp, defines fiducials and head surface points. Coordinate labels are included in the file, so no sfn file is required.
- The cot file, MEGASCII.cot, defines the coordinates of the head center. If this were missing, BESA Research would compute the head center based on the sphere that best fits the head surface points.
- The pos file, MEGASCII.pos, defines the coordinates of the 122 sensors. For the Neuromag system there are 9 values per line, defining primary coil location, secondary coil location, and orientation cosines. The sequence of coordinates in the pos file must match the sequence of MEG channels! The file format and locations of the primary and secondary coils allow BESA Research to identify the sensor type as planar gradiometers. If the file had only six values per line, BESA Research would classify the sensors as magnetometers (one primary coil and the orientation cosines).
- Open the file, selecting current file type as ‘*,m??’. The Channel and digitized head surface point information dialog box will open, displaying the different auxiliary file names. The green tick indicates that BESA Research finds everything to be OK.
- Press the ‘OK’ button and then the ‘V’ key to view the coordinates.
Example 2. File Import
- The file, MEGASCIIimport.asc, contains MEG data in a multiplexed array, without a header. This needs to be imported using File/Import ASCII (see “Example: ASCII Import”).
- On import you have to specify the file as ‘Multiplexed’, the number of time points (285), the number of channels (132), the bins/µV (or bins/fT) (=1), the time at which the stimulus occurred (50 ms), and the sampling rate (949.667 Hz).
- This format contains no channel labels. The labels in MEGASCIIimport.ela are recognized by BESA Research as originating from the Neuromag system. They are therefore identified as MEG and do not need further identification.
- Since it recognizes the channels as MEG, the Channel and digitized head surface point information dialog box will open, displaying the different auxiliary file names as before. Since all necessary files with the same base name as the data file are supplied, they are read automatically.
- Press the ‘OK’ button, enter a segment name, and then the ‘V’ key to view the coordinates.
Example 3. File Open -- MEG information recorded elsewhere
This example illustrates the case where the auxiliary files have a different base name from the data file: you must select the file name in the Channel and digitized head surface point information dialog box.
- Open the file, MEGASCIIelsewhere.mul. It is read as an MEG magnetometer file.
- In the Channel and digitized head surface point information dialog box, specify
- MEG coordinates will be correct. The sensor definition file specifies the sensors as planar gradiometers.
- Where the auxiliary files came from will be recorded in the database. If you open the file again, the auxiliary files will be found automatically, without asking any questions.
Example: Reading combined EEG and MEG from an ASCII file
The files described in this example can be found in the Examples\Xtras\MEG+EEG subdirectory of the BESA Research installation folder.
Here are two examples containing mixed MEG, EEG, and polygraphic channels:
- Open a file using the File/Open command
- Import a file using File/Import ASCII command
In both cases
- The *.ela file defines the channel labels. Based on the labels, BESA Research knows which channels are EEG and MEG. The remainder are classified as polygraphic channels.
- The *.pos file defines the MEG sensor coordinates. The number of values on a line of this file (=9) defines the MEG as gradiometers. The relative locations of primary and secondary coils identify the gradiometers as planar.
1. Example with File/Open
- Open the file MEG+EEG.mul. The Channel and digitized head surface point information dialog box will open automatically (unless the file has already been read once and the information is in the database).
- You will see under ‘internal data file information’ that BESA Research finds 122 MEG sensors, and 162 channels in all.
- Under ‘Channel configuration’, you will see that as a result of reading the file MEG+EEG.ela, 32 channels are defined as EEG, and 8 channels as polygraphic.
- Under ‘Digitized head surface points’ is the feedback that out of the 51 locations in the file MEG+EEG.sfp, all electrode locations have been defined.
- Under ‘MEG sensors’, the sensors have been identified as gradiometers.
- The green tick at the top right of the window indicates that BESA Research classifies everything as OK.
2. Example with File/Import ASCII
- Import the file MEG+EEGimport.asc. Select MEG+EEGimport.fsg as the target file (see “Example: ASCII Import”).
- Select 320 Hz sampling rate, and 500 ms pre-stimulus time. Other selections in the dialog box should be ‘Multiplexed’, 1 bin/microvolt (this is interpreted as 1 bin/fT for MEG), 162 channels and 320 samples.
- The Channel and digitized head surface point information dialog box will open as above.