Difference between revisions of "Export Single Trial Data"
| (19 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
| + | {{BESAInfobox | ||
| + | |title = Module information | ||
| + | |module = BESA Research Basic or higher | ||
| + | |version = 5.2 or higher | ||
| + | }} | ||
| + | |||
== Rationale == | == Rationale == | ||
This article describes how to export single-trial EEG or MEG raw data for further analysis in an external program, e.g. in Matlab. | This article describes how to export single-trial EEG or MEG raw data for further analysis in an external program, e.g. in Matlab. | ||
| + | |||
== Steps to follow == | == Steps to follow == | ||
| − | === | + | |
| + | === Load data in BESA === | ||
| + | |||
The data are then displayed in the review window. Now you can either | The data are then displayed in the review window. Now you can either | ||
* use the ERP module to define a condition and reject any trials with artifacts (see the BESA Help section on ERP for more information), or | * use the ERP module to define a condition and reject any trials with artifacts (see the BESA Help section on ERP for more information), or | ||
* directly move to the export of all epochs around a particular trigger. | * directly move to the export of all epochs around a particular trigger. | ||
| − | |||
| − | [[File:Export Epochs | + | === Export data from BESA === |
| + | |||
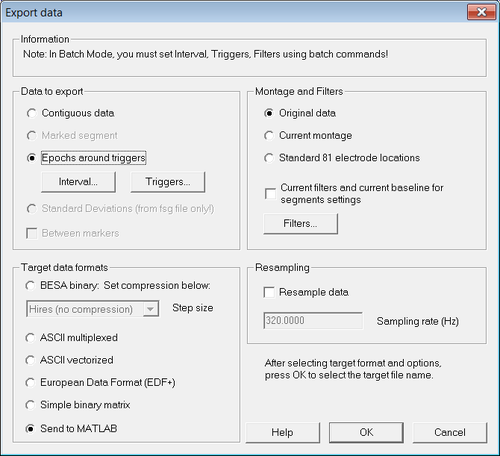
| + | Select "''File → Export...''". The [[Export#Export Dialog|export dialog]] opens: | ||
| + | |||
| + | [[File:Export_epochs_1.png|500px|thumb|c|none|Figure 1 "Export data" dialog]] | ||
| + | |||
| + | |||
| + | Make sure that the options "Epochs around triggers" and "Original data" are selected. In the section "Target formats", choose the option of choice: | ||
| + | * If you want to create ASCII files for offline analysis, select "[[ASCII File Format#ASCII Multiplexed file format|ASCII multiplexed]]" or "[[ASCII File Format#ASCII Vectorized file format|ASCII vectorized]]" | ||
| + | * If you want to analyze data directly in Matlab, select "[[#Data in MATLAB|Send to MATLAB]]" | ||
| + | |||
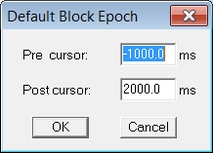
| + | For the "''Interval...''" select a long enough time span as shown below: | ||
| + | |||
| + | [[File:Export_epochs_2.png|213px|thumb|c|none|Figure 2 "Epoch around trigger" dialog]] | ||
| + | |||
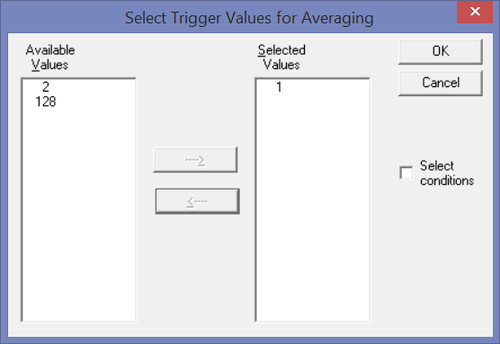
| + | Then click the "''Triggers...''" button to select which trigger code or condition will be used. | ||
| + | |||
| + | If you want to export the epochs around a trigger code: Make sure that the trigger code you want is selected - e.g. the trigger code 1 in this example. | ||
| + | |||
| + | [[File:Export_epochs_3.png|500px|thumb|c|none|Figure 3 "Trigger codes" dialog]] | ||
| + | |||
| + | |||
| + | If you want to export the epochs around a condition: Check the box "Conditions". Then ensure that the condition of choice is selected (e.g. condition "Rare" in the example in Figure 4). | ||
| + | |||
| + | [[File:Export_epochs_4.png|500px|thumb|c|none|Figure 4 "Epoch around condition" dialog]] | ||
| + | |||
| + | |||
| + | After clicking "OK" BESA Research either exports the data, or - if that option was chosen - sends the data to Matlab. | ||
| + | In case of Matlab, it opens the Matlab window and creates the structure "besa_channels". From Matlab save the structure as "besa_channels.mat". | ||
| + | |||
| + | === Data in MATLAB === | ||
| + | |||
| + | The MATLAB structure <code>besa_channels</code> contains several fields like in the following example of a 64-channel EEG file with 39 trials that survived the artifact scan: | ||
| + | |||
| + | <source lang="matlab"> | ||
| + | datafile: [1x93 char] | ||
| + | channeltypes: {1x64 cell} | ||
| + | channellabels: {1x64 cell} | ||
| + | channelunits: {1x64 cell} | ||
| + | channelcoordinates: [64x9 double] | ||
| + | montage: 'Original' | ||
| + | filters: 'Filters off' | ||
| + | samplingrate: 100 | ||
| + | HSPcoordinates: [3x3 double] | ||
| + | HSPtypes: {'Fiducial' 'Fiducial' 'Fiducial'} | ||
| + | HSPlabels: {'FidNz' 'FidT9' 'FidT10'} | ||
| + | headcenter: [-0.0020 8.1247e-004 0.0291] | ||
| + | headradius: 0.1021 | ||
| + | electrodethickness: 0 | ||
| + | structtype: 'besa_channels' | ||
| + | datatype: 'Epoched_Data' | ||
| + | data: [1x39 struct] | ||
| + | </source > | ||
| + | |||
| + | The sub-structure data contains the latencies and the measured amplitudes for each channel and trial: | ||
| + | |||
| + | <source lang="dos"> | ||
| + | >> besa_channels.data | ||
| + | |||
| + | ans = | ||
| + | |||
| + | 1x39 struct array with fields: | ||
| + | amplitudes | ||
| + | timeoffsetsecs | ||
| + | latencies | ||
| + | event | ||
| + | </source > | ||
| + | |||
| + | [[Category:ERP/ERF]] [[Category:Data Import/Export]] | ||
Latest revision as of 15:53, 5 May 2021
| Module information | |
| Modules | BESA Research Basic or higher |
| Version | 5.2 or higher |
Contents
Rationale
This article describes how to export single-trial EEG or MEG raw data for further analysis in an external program, e.g. in Matlab.
Steps to follow
Load data in BESA
The data are then displayed in the review window. Now you can either
- use the ERP module to define a condition and reject any trials with artifacts (see the BESA Help section on ERP for more information), or
- directly move to the export of all epochs around a particular trigger.
Export data from BESA
Select "File → Export...". The export dialog opens:
Make sure that the options "Epochs around triggers" and "Original data" are selected. In the section "Target formats", choose the option of choice:
- If you want to create ASCII files for offline analysis, select "ASCII multiplexed" or "ASCII vectorized"
- If you want to analyze data directly in Matlab, select "Send to MATLAB"
For the "Interval..." select a long enough time span as shown below:
Then click the "Triggers..." button to select which trigger code or condition will be used.
If you want to export the epochs around a trigger code: Make sure that the trigger code you want is selected - e.g. the trigger code 1 in this example.
If you want to export the epochs around a condition: Check the box "Conditions". Then ensure that the condition of choice is selected (e.g. condition "Rare" in the example in Figure 4).
After clicking "OK" BESA Research either exports the data, or - if that option was chosen - sends the data to Matlab.
In case of Matlab, it opens the Matlab window and creates the structure "besa_channels". From Matlab save the structure as "besa_channels.mat".
Data in MATLAB
The MATLAB structure besa_channels contains several fields like in the following example of a 64-channel EEG file with 39 trials that survived the artifact scan:
datafile: [1x93 char] channeltypes: {1x64 cell} channellabels: {1x64 cell} channelunits: {1x64 cell} channelcoordinates: [64x9 double] montage: 'Original' filters: 'Filters off' samplingrate: 100 HSPcoordinates: [3x3 double] HSPtypes: {'Fiducial' 'Fiducial' 'Fiducial'} HSPlabels: {'FidNz' 'FidT9' 'FidT10'} headcenter: [-0.0020 8.1247e-004 0.0291] headradius: 0.1021 electrodethickness: 0 structtype: 'besa_channels' datatype: 'Epoched_Data' data: [1x39 struct]
The sub-structure data contains the latencies and the measured amplitudes for each channel and trial:
>> besa_channels.data
ans =
1x39 struct array with fields:
amplitudes
timeoffsetsecs
latencies
event