Difference between revisions of "How to Use BrainVision Analyzer with BESA Connectivity"
(Created page with "{{BESAInfobox |title = Module information |module = BESA Connectivity |version = BESA Connectivity 1.0 or higher }} == Note and general remarks== To transform data from Brai...") |
|||
| (2 intermediate revisions by the same user not shown) | |||
| Line 7: | Line 7: | ||
== Note and general remarks== | == Note and general remarks== | ||
| − | To transform data from BrainVision Analyzer you need to convert it using MATLAB. We assume that you have it already installed on your PC and BrainVision Analyzer is configured to work with it. You will also need this script from our | + | To transform data from BrainVision Analyzer to BESA Connectivity, you need to convert it using MATLAB. We assume that you have it already installed on your PC and BrainVision Analyzer is configured to work with it. You will also need this script from our Github page: |
| + | |||
https://github.com/BESA-GmbH/BESA-MATLAB-Scripts/blob/main/Additional_utilities/BESAConnectivityScripts/Analyzer_to_BESAConnectivity.m | https://github.com/BESA-GmbH/BESA-MATLAB-Scripts/blob/main/Additional_utilities/BESAConnectivityScripts/Analyzer_to_BESAConnectivity.m | ||
You will also need these toolboxes: | You will also need these toolboxes: | ||
https://github.com/BESA-GmbH/BESA-MATLAB-Scripts/tree/main/MATLAB2BESA | https://github.com/BESA-GmbH/BESA-MATLAB-Scripts/tree/main/MATLAB2BESA | ||
| + | |||
| + | Please ensure that the MATLAB2BESA toolbox is in the MATLAB path. | ||
== What to do in BrainVision Analyzer== | == What to do in BrainVision Analyzer== | ||
| − | You need to prepare data that is in segments | + | You need to prepare data that is in segments - but not averaged! You can of course apply any filtering and data processing beforehand. Then you export segments to MATLAB using the Brain Vision Analyzer to MATLAB interface. |
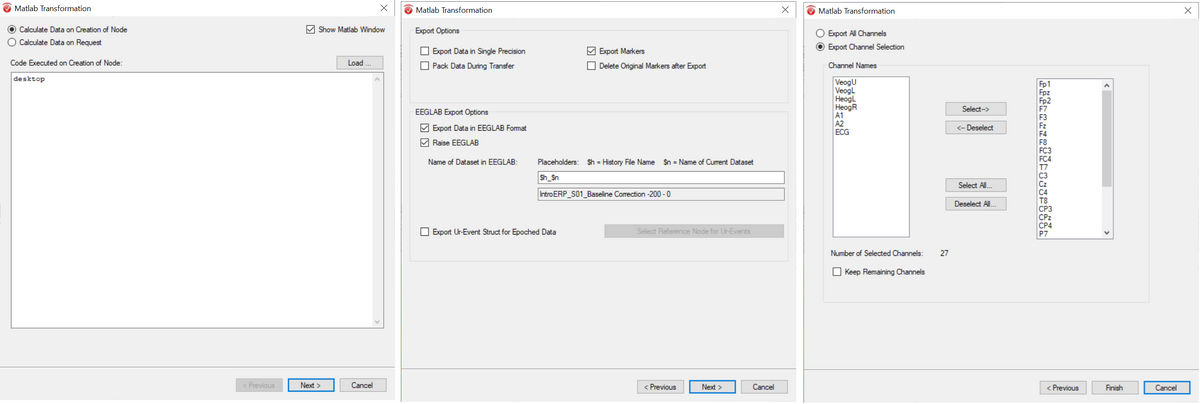
| + | Please make sure to use the following options when exporting: | ||
| + | - Type "desktop" in the first dialog | ||
| + | - Check boxes for "Export data in EEGLab format", "Raise EEGLab" and "Markers" in the second dialog. | ||
| + | - In the third dialog, select all EEG channels that have coordinates (but not channels which do not have coordinates in Brain Vision Analyzer, like A1, A2) | ||
| + | |||
| + | |||
| + | [[File:Analyzer Export Options.png|1200px]] | ||
== Adapt the script == | == Adapt the script == | ||
| − | + | The script Analyzer_to_BESAConnectivity.m needs to be adapted. The following lines should be changed: | |
*In line after "%% Add toolboxes" provide full path to MATLAB2BESA toolbox | *In line after "%% Add toolboxes" provide full path to MATLAB2BESA toolbox | ||
*You may adapt where the resulting file will be saved by changing the line "FilePathName = [pwd '\BAtoBESA.generic'];" | *You may adapt where the resulting file will be saved by changing the line "FilePathName = [pwd '\BAtoBESA.generic'];" | ||
| + | *There may be a different convention for x-axis values in the EEGLab representation. If electrodes like F4, P4, etc. appear on the left, then adjust the script such that in section ''Channel labels and units'' of the script, the line | ||
| + | : <code>EEG.chanlocs(ChanIdx).Y,... </code> is replaced by | ||
| + | : <code>-EEG.chanlocs(ChanIdx).Y,... </code> | ||
== Finalization == | == Finalization == | ||
You just need to run the script just by either pressing Run in matlab toolbar or by calling "Analyzer_to_BESAConnectivity" command. | You just need to run the script just by either pressing Run in matlab toolbar or by calling "Analyzer_to_BESAConnectivity" command. | ||
| − | In BESA Connectivity you open the newly created file. | + | In BESA Connectivity you then start a Time-Frequency workflow and open the newly created file. |
[[Category:Connectivity]] [[Category:Time-Frequency]] [[Category:Data Import/Export]] | [[Category:Connectivity]] [[Category:Time-Frequency]] [[Category:Data Import/Export]] | ||
Latest revision as of 20:27, 15 November 2023
| Module information | |
| Modules | BESA Connectivity |
| Version | BESA Connectivity 1.0 or higher |
Contents
Note and general remarks
To transform data from BrainVision Analyzer to BESA Connectivity, you need to convert it using MATLAB. We assume that you have it already installed on your PC and BrainVision Analyzer is configured to work with it. You will also need this script from our Github page:
https://github.com/BESA-GmbH/BESA-MATLAB-Scripts/blob/main/Additional_utilities/BESAConnectivityScripts/Analyzer_to_BESAConnectivity.m You will also need these toolboxes: https://github.com/BESA-GmbH/BESA-MATLAB-Scripts/tree/main/MATLAB2BESA
Please ensure that the MATLAB2BESA toolbox is in the MATLAB path.
What to do in BrainVision Analyzer
You need to prepare data that is in segments - but not averaged! You can of course apply any filtering and data processing beforehand. Then you export segments to MATLAB using the Brain Vision Analyzer to MATLAB interface. Please make sure to use the following options when exporting: - Type "desktop" in the first dialog - Check boxes for "Export data in EEGLab format", "Raise EEGLab" and "Markers" in the second dialog. - In the third dialog, select all EEG channels that have coordinates (but not channels which do not have coordinates in Brain Vision Analyzer, like A1, A2)
Adapt the script
The script Analyzer_to_BESAConnectivity.m needs to be adapted. The following lines should be changed:
- In line after "%% Add toolboxes" provide full path to MATLAB2BESA toolbox
- You may adapt where the resulting file will be saved by changing the line "FilePathName = [pwd '\BAtoBESA.generic'];"
- There may be a different convention for x-axis values in the EEGLab representation. If electrodes like F4, P4, etc. appear on the left, then adjust the script such that in section Channel labels and units of the script, the line
-
EEG.chanlocs(ChanIdx).Y,...is replaced by -
-EEG.chanlocs(ChanIdx).Y,...
Finalization
You just need to run the script just by either pressing Run in matlab toolbar or by calling "Analyzer_to_BESAConnectivity" command. In BESA Connectivity you then start a Time-Frequency workflow and open the newly created file.