Difference between revisions of "Seed dipole solutions from cortical images"
| (6 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
{{BESAInfobox | {{BESAInfobox | ||
|title = Module information | |title = Module information | ||
| − | |module = BESA Research | + | |module = BESA Research Standard or higher |
|version = 5.2 or higher | |version = 5.2 or higher | ||
}} | }} | ||
==Overview== | ==Overview== | ||
| − | BESA initially was designed to compute comprehensive dipole | + | BESA initially was designed to compute comprehensive dipole solutions using so-called inverse methods. With the advance of neuroinformatics we also incorporated many popular volumetric solutions (e.g. LORETA) and our own algorithms (e.g. CLARA). Since the release of BESA Research 6.1 cortical solutions (e.g. Cortical LORETA) are also available. |
| − | As always both inverse problem | + | As always both inverse problem solution approaches (dipole and 3D Imaging methods) have some pros and cons. For example, for dipole analysis, the number of dipoles has to be assumed by the user. 3D Imaging methods do not need such assumptions, but need additional constraints such as regularization in order to obtain a solution for the inverse problem. Also, the orientation and wave form of the sources in the (cortical) image solution is not readily available. |
| − | + | To overcome these limitations, there is the possibility to seed dipole(s) from 3D and cortical images. | |
| − | To overcome | + | |
==Seeding dipoles== | ==Seeding dipoles== | ||
| − | When you have a | + | When you have a Source Analysis window open and you used a 3D Imaging source analysis of your choice you might end up with something similar to this (the CLARA algorithm was used in this example): |
[[File:DipoleFromSourceArrows.jpg|800px]] | [[File:DipoleFromSourceArrows.jpg|800px]] | ||
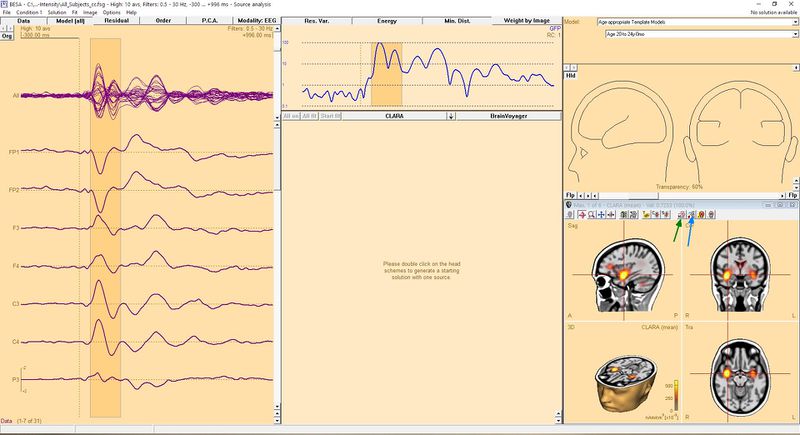
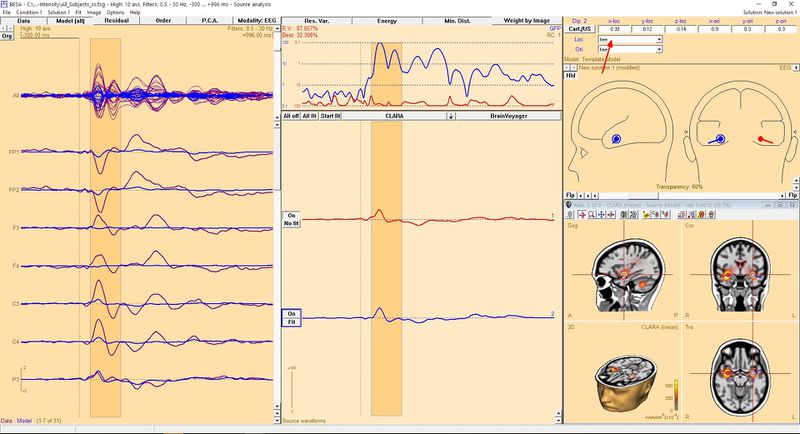
| − | To seed a dipole in exact place where the source is, | + | To seed a dipole in the exact place where the source is, click the toolbar button named "Switch to next grid maximum" (the button with the small 'M' letter, indicated by a green arrow in the image). This will place a crosshair in the exact place of the source maximum designated by the 3D image algorithm. You can click this button once again if you want to switch to the next maximum. If you press the button while holding the Ctrl key, you go back to the previous maximum. When you find the place where you want to seed the dipole, just click the button with the dipole symbol on it (indicated with a blue arrow in the Figure above). Of course you can repeat these steps to seed more than one dipole. For example, in the presented case you might want to seed two dipoles, as CLARA indicates two potential sources (in both hemispheres in the temporal lobes). After doing this we get the following result: |
[[File:DipoleFromSource2arrow.jpg|800px]] | [[File:DipoleFromSource2arrow.jpg|800px]] | ||
==Fitting dipoles== | ==Fitting dipoles== | ||
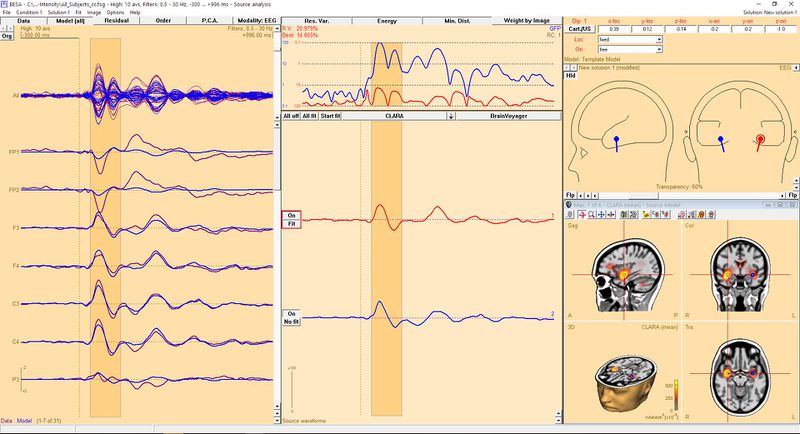
| − | To get better approximation of | + | To get a better approximation of dipole locations and orientations, we should fit them. Since the dipoles were placed on the basis of 3D image, a good idea is to keep their location in the original position. So, for each dipole it is advisable to change the location from "free" to "fixed" by selecting this entry from the list indicated by the red arrow in the image above. You can switch between dipoles just by clicking on them. After this is done you can fit all dipoles to the data. Click the "All fit" button in the central window to select all dipoles for fitting. Now press "Start fit". The result of this operation is presented below. Please note that the waveforms and orientations have changed but location has not. |
| + | [[File:DipoleFromSourceFinal.JPG|800px]] | ||
| − | + | ==Dipole seeding from cortical solution example== | |
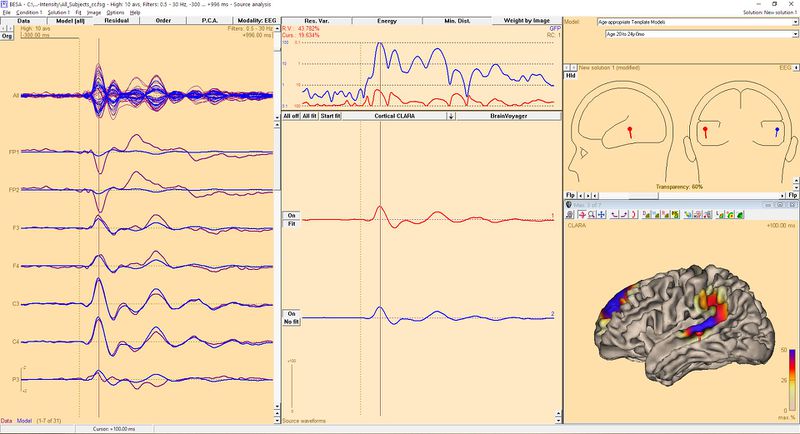
| + | As mentioned above, dipoles can be seeded also from cortical solutions (see example below). This approach allows evaluating sources very precisely. Because of brain physiology and the neocortex structure dipoles would be expected to be oriented approximately perpendicular to the brain surface. | ||
| + | [[File:DipoleFromSourceCortical.JPG|800px]] | ||
[[Category:Source Analysis]] | [[Category:Source Analysis]] | ||
Latest revision as of 16:37, 20 February 2017
| Module information | |
| Modules | BESA Research Standard or higher |
| Version | 5.2 or higher |
Contents
Overview
BESA initially was designed to compute comprehensive dipole solutions using so-called inverse methods. With the advance of neuroinformatics we also incorporated many popular volumetric solutions (e.g. LORETA) and our own algorithms (e.g. CLARA). Since the release of BESA Research 6.1 cortical solutions (e.g. Cortical LORETA) are also available. As always both inverse problem solution approaches (dipole and 3D Imaging methods) have some pros and cons. For example, for dipole analysis, the number of dipoles has to be assumed by the user. 3D Imaging methods do not need such assumptions, but need additional constraints such as regularization in order to obtain a solution for the inverse problem. Also, the orientation and wave form of the sources in the (cortical) image solution is not readily available. To overcome these limitations, there is the possibility to seed dipole(s) from 3D and cortical images.
Seeding dipoles
When you have a Source Analysis window open and you used a 3D Imaging source analysis of your choice you might end up with something similar to this (the CLARA algorithm was used in this example):
To seed a dipole in the exact place where the source is, click the toolbar button named "Switch to next grid maximum" (the button with the small 'M' letter, indicated by a green arrow in the image). This will place a crosshair in the exact place of the source maximum designated by the 3D image algorithm. You can click this button once again if you want to switch to the next maximum. If you press the button while holding the Ctrl key, you go back to the previous maximum. When you find the place where you want to seed the dipole, just click the button with the dipole symbol on it (indicated with a blue arrow in the Figure above). Of course you can repeat these steps to seed more than one dipole. For example, in the presented case you might want to seed two dipoles, as CLARA indicates two potential sources (in both hemispheres in the temporal lobes). After doing this we get the following result:
Fitting dipoles
To get a better approximation of dipole locations and orientations, we should fit them. Since the dipoles were placed on the basis of 3D image, a good idea is to keep their location in the original position. So, for each dipole it is advisable to change the location from "free" to "fixed" by selecting this entry from the list indicated by the red arrow in the image above. You can switch between dipoles just by clicking on them. After this is done you can fit all dipoles to the data. Click the "All fit" button in the central window to select all dipoles for fitting. Now press "Start fit". The result of this operation is presented below. Please note that the waveforms and orientations have changed but location has not.
Dipole seeding from cortical solution example
As mentioned above, dipoles can be seeded also from cortical solutions (see example below). This approach allows evaluating sources very precisely. Because of brain physiology and the neocortex structure dipoles would be expected to be oriented approximately perpendicular to the brain surface.