Difference between revisions of "Export Single Trial Data"
| Line 31: | Line 31: | ||
After clicking "OK" BESA Research either exports the data, or - if that option was chosen - sends the data to Matlab. | After clicking "OK" BESA Research either exports the data, or - if that option was chosen - sends the data to Matlab. | ||
In case of Matlab, it opens the Matlab window and creates the structure "besa_channels". From Matlab save the structure as "besa_channels.mat". | In case of Matlab, it opens the Matlab window and creates the structure "besa_channels". From Matlab save the structure as "besa_channels.mat". | ||
| − | === 3 | + | === 3 Data in Matlab === |
| − | The Matlab structure "besa_channels" contains several fields like in the following example of a | + | The Matlab structure "besa_channels" contains several fields like in the following example of a 64-channel EEG file with 39 trials that survived the artifact scan: |
<tt> | <tt> | ||
| − | datafile: [ | + | datafile: [1x93 char] |
| − | channeltypes: { | + | channeltypes: {1x64 cell} |
| − | channellabels: { | + | channellabels: {1x64 cell} |
| − | channelunits: { | + | channelunits: {1x64 cell} |
| − | channelcoordinates: [ | + | channelcoordinates: [64x9 double] |
montage: 'Original' | montage: 'Original' | ||
filters: 'Filters off' | filters: 'Filters off' | ||
| − | samplingrate: | + | samplingrate: 100 |
HSPcoordinates: [3x3 double] | HSPcoordinates: [3x3 double] | ||
HSPtypes: {'Fiducial' 'Fiducial' 'Fiducial'} | HSPtypes: {'Fiducial' 'Fiducial' 'Fiducial'} | ||
HSPlabels: {'FidNz' 'FidT9' 'FidT10'} | HSPlabels: {'FidNz' 'FidT9' 'FidT10'} | ||
| − | headcenter: [ | + | headcenter: [-0.0020 8.1247e-004 0.0291] |
| − | headradius: 0. | + | headradius: 0.1021 |
electrodethickness: 0 | electrodethickness: 0 | ||
structtype: 'besa_channels' | structtype: 'besa_channels' | ||
datatype: 'Epoched_Data' | datatype: 'Epoched_Data' | ||
| − | data: [ | + | data: [1x39 struct] |
| + | </tt> | ||
| + | |||
| + | The sub-structure data contains the latencies and the measured amplitudes for each channel and trial: | ||
| + | <tt> | ||
| + | » besa_channels.data | ||
| + | |||
| + | ans = | ||
| + | |||
| + | 1x39 struct array with fields: | ||
| + | amplitudes | ||
| + | timeoffsetsecs | ||
| + | latencies | ||
| + | event | ||
</tt> | </tt> | ||
Revision as of 16:13, 7 March 2016
Contents
Rationale
This article describes how to export single-trial EEG or MEG raw data for further analysis in an external program, e.g. in Matlab.
Steps to follow
1 Load data in BESA
The data are then displayed in the review window. Now you can either
- use the ERP module to define a condition and reject any trials with artifacts (see the BESA Help section on ERP for more information), or
- directly move to the export of all epochs around a particular trigger.
2 Export data from BESA
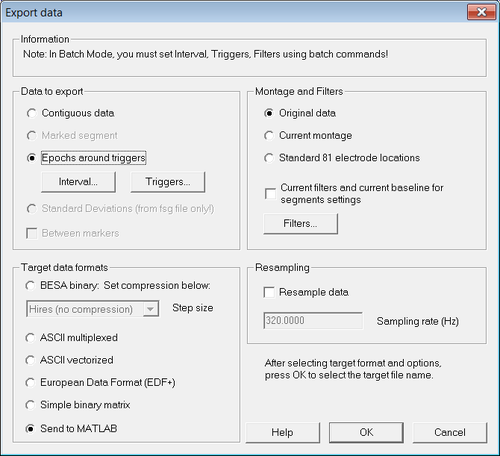
Select "File -> Export...". The export dialog opens:
Make sure that the options "Epochs around triggers" and "Original data" are selected. In the section "Target formats", choose the option of choice:
- If you want to create ASCII files for offline analysis, select "ASCII multiplexed" or "ASCII vectorized"
- If you want to analyze data directly in Matlab, select "Send to MATLAB"
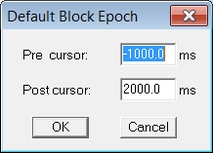
For the "Interval..." select a long enough time span as shown in figure 2:
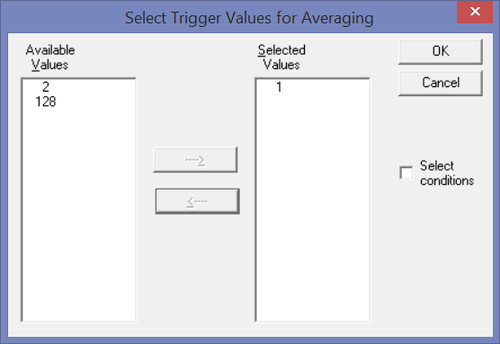
Then click the "Triggers..." button to select which trigger code or condition will be used. If you want to export the epochs around a trigger code: Make sure that the trigger code you want is selected - e.g. the trigger code 1 in this example.
If you want to export the epochs around a condition: Check the box "Conditions". Then ensure that the condition of choice is selected (e.g. condition "Rare" in the example.
After clicking "OK" BESA Research either exports the data, or - if that option was chosen - sends the data to Matlab. In case of Matlab, it opens the Matlab window and creates the structure "besa_channels". From Matlab save the structure as "besa_channels.mat".
3 Data in Matlab
The Matlab structure "besa_channels" contains several fields like in the following example of a 64-channel EEG file with 39 trials that survived the artifact scan:
datafile: [1x93 char]
channeltypes: {1x64 cell}
channellabels: {1x64 cell}
channelunits: {1x64 cell}
channelcoordinates: [64x9 double]
montage: 'Original'
filters: 'Filters off'
samplingrate: 100
HSPcoordinates: [3x3 double]
HSPtypes: {'Fiducial' 'Fiducial' 'Fiducial'}
HSPlabels: {'FidNz' 'FidT9' 'FidT10'}
headcenter: [-0.0020 8.1247e-004 0.0291]
headradius: 0.1021
electrodethickness: 0
structtype: 'besa_channels'
datatype: 'Epoched_Data'
data: [1x39 struct]
The sub-structure data contains the latencies and the measured amplitudes for each channel and trial: » besa_channels.data
ans =
1x39 struct array with fields:
amplitudes timeoffsetsecs latencies event