Useful batches
| Module information | |
| Modules | BESA Research Standard or higher |
| Version | 7.1 or higher |
This page lists a number of useful batches that you can try yourself.
Contents
Download the batch files
Download and unzip the following files into your folder C:\Users\Public\Documents\BESA\Research_7_1\Scripts\Batch\ (you may need to replace C:\Users\Public\Documents\ with the equivalent Public Documents folder on your computer).
Then move the file TenDipBlue.bsa to the folder C:\Users\Public\Documents\BESA\Research_7_1\Scripts\ColorSchemes\ (you may need to replace C:\Users\Public\Documents\ with the equivalent Public Documents folder on your computer).
ECG correction
Batch name
ECGCorrection.bbat
What does it do?
This batch is used for correcting ECG artifacts. It is particularly useful for MEG data. It should be called before creating an MEG source montage.
How to use it
Load a data set that contains an ECG channel, or a clear manifestation of the ECG artifact. Make sure that the channel label of this channel is visible at the left (if not, adjust the montage, or change displayed channel types using the buttons at the top right, until you can see it). Run the batch. The batch will ask you to set a cursor on the peak of the signal, and mark the channel if possible. It will then mark a number of occurrences and use tag #2 to collect the average ECG signal. This will then be used as an artifact topography for an adaptive artifact correction with minimum distortion of signal of interest.
Create an MEG source montage
Batch name
CreateMEGSourceMtg.bbat
CreateGRDSourceMtg.bbat
What does it do?
This batch is used for creating an MEG source montage, taking an existing artifact correction for ECG artifacts into account. The batch creates a magnetometer-based [gradiometer-based] source montage with 29 montage channels. Each channel has two orientations. Thus, a maximum of 58 source waveforms will be displayed. The waveforms depict the signal originating from the brain region underlying the head position indicated by the channel label (e.g. F3L will be a brain region underneath the location of the F3 channel from the 10-10 nomenclature).
This batch will enable you to use the MEG review as described in the publications by Benicky et al. (2017)[1] and Nenonen et al. (2022) [2].
How to use it
It is recommended to first run ECGCorrection.bbat, or create an ECG artifact topography manually. Then run the batch. The batch CreateMEGSourceMtg creates a 29-channel source montage based on magnetometers or axial gradiometers. In case that planar gradiometers should be used, please use the batch CreateGRDSourceMtg.bbat. After finalization, the number of source waveforms displayed can be adjusted using the button “Opt” – either use Regional Source oriented, or Regional Source all.
Moving dipole fit
Batch name
MovingDipole11Dips_Minus20ToPeak.bbat
Additional file: TenDipBlue.bsa
What does it do?
This batch is intended to be used on epileptic spike averages. It performs a single dipole fit for each time point between 20 ms before the spike peak to the peak, in 2 ms steps. It displays the result in the 3D MRI view of the subject.
How to use it
As a pre-requisite, the associated solution file TenDipBlue.bsa needs to be copied to the folder C:\Users\Public\Documents\BESA\Research_7_1\Scripts\ColorSchemes.
An averaged segment containing an epileptic spike should be loaded. It is assumed that the spike maximum lies at the zero latency point of the segment (indicated by a dotted line in the review display).
Before running the batch, check the filter settings: For epileptic spikes, we recommend setting a low cutoff of 3 Hz forward filter with 6dB, and a high cutoff of 35 Hz zero-phase with 24 dB.
Run the batch and follow the instructions when prompted. The result is automatically saved in a solution file (file basename followed by _MovDip_minus20_to_peak.bsa).
Dipole cluster fit
Batch name
ClusterDipoleFit.bbat
What does it do?
For a number of segments of single events, the batch performs a dipole fit in each segment, and displays the result as a cluster in the 3D MRI view of the subject.
How to use it
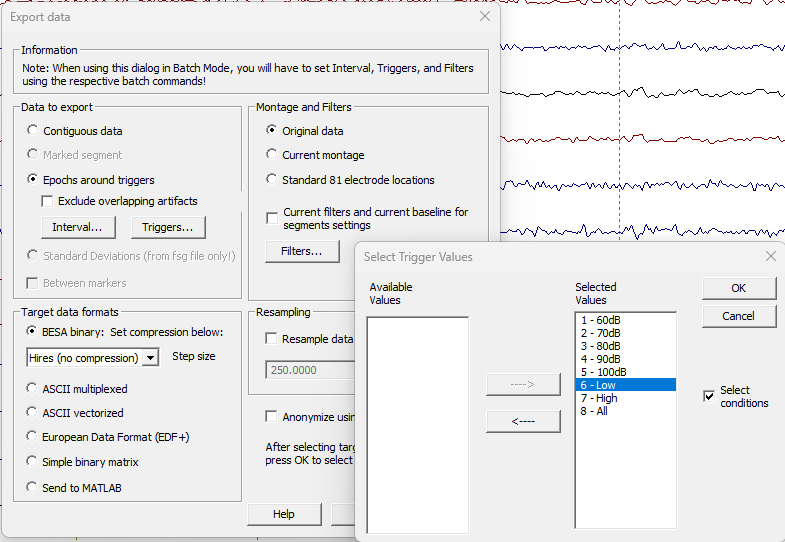
As a prerequisite, you will need a segmented file that contains segments of single events of equal size. In order to get this, you can use the menu “File / Export”, then select “Epochs around triggers”, then press the button “Triggers” where you can define which trigger(s) to use (you can also use conditions that you defined in the ERP module).
The interval can be adjusted with the “Interval” button. The batch will expect at least 100 ms before and more than 100 ms after the event time point.
Run the export, and re-load the exported file.
Before running the batch, check the filter settings: For epileptic spikes, we recommend setting a low cutoff of 3 Hz forward filter with 6dB, and a high cutoff of 35 Hz zero-phase with 24 dB. Also, in case you have both MEG and EEG data in your file, select the general modality you want to fit (EEG or MEG) using the button at the top right of the review window.
Now, start the batch. You will be asked to supply the latency at which the fit should be performed. Then, all segments will be processed (up to a maximum of 50 segments).
Create discrete solution from source image
Batch name
CreateSourcesFromImageEEG.bbat
CreateSourcesFromImageMEG.bbat
What does it do?
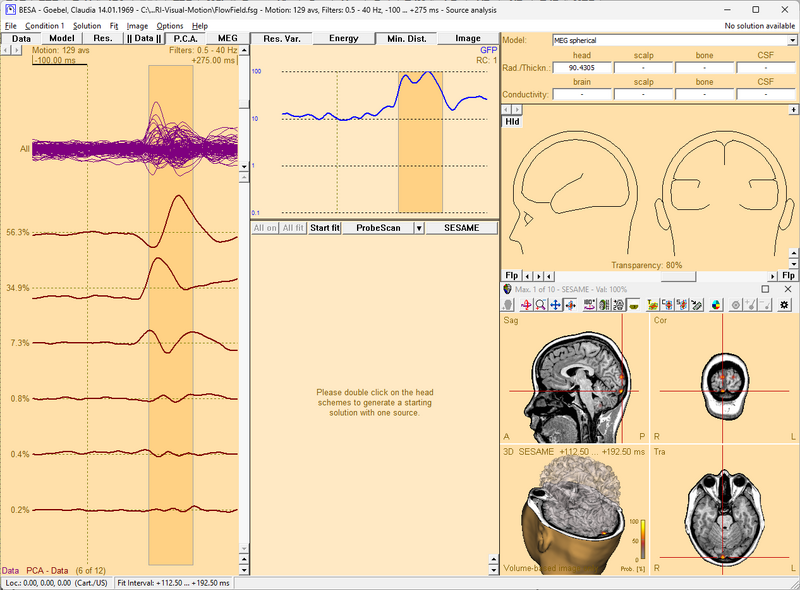
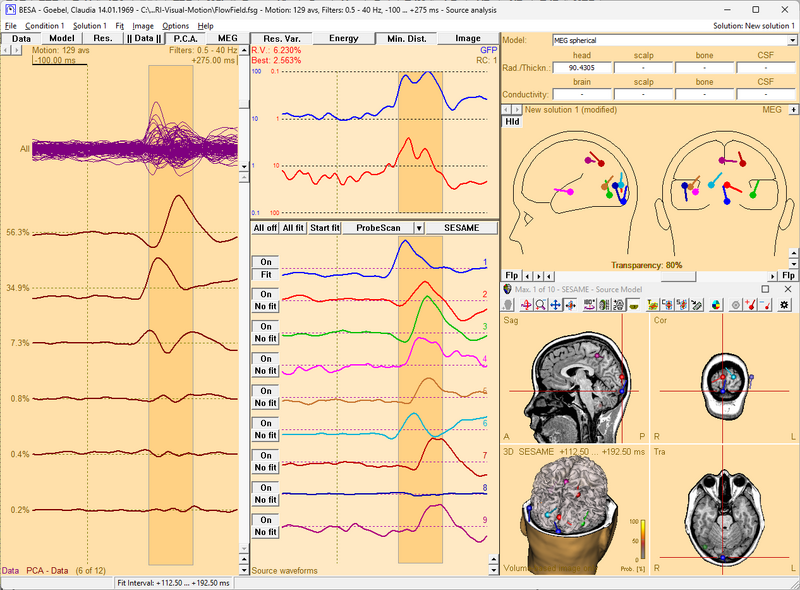
After a source image has been computed, this batch (with EEG in the name for an EEG image, or with MEG in the name for an MEG image) will create a new discrete solution, add the sources from the maxima of the source image, and fit their orientation within the current fit interval.
How to use it
As a prerequisite, you will need to compute a source image in the Source Analysis window. The batch should then be called directly from the BESA Source Analysis window, using the menu Fit / Run Batch. It will ask for the number of maxima in the image, which you can read from the title bar of the 3D window. Then it will create the solution and ensure that orientations are fitted correctly.