Difference between revisions of "Integration of Custom Template Head-Models"
(→Coregistration of EEG and MRI Data) |
(→Create new Folder for Custom Template Head-Model) |
||
| Line 42: | Line 42: | ||
=== Create new Folder for Custom Template Head-Model === | === Create new Folder for Custom Template Head-Model === | ||
| + | |||
| + | To add a new template head-model for BESA Research, navigate to the installation directory. The default installation directory for BESA Research 6.1 is: | ||
| + | |||
| + | <source lang="dos"> | ||
| + | C:\Program Files (x86)\BESA\Research_6_1\ | ||
| + | </source > | ||
| + | |||
| + | and create a new folder (e.g. ''0y3mo'') in the ''.\System\TemplateHeadModels\AgeAppropriateTemplates\'' subfolder. | ||
=== Copy Files to Custom Template Head-Model Folder === | === Copy Files to Custom Template Head-Model Folder === | ||
Revision as of 12:59, 30 November 2017
| Module information | |
| Modules | BESA Research, BESA MRI |
| Version | 6.1 or higher, 2.0 or higher |
Contents
- 1 Introduction
- 2 Prerequisites
- 3 Custom Template Head-Model Integration in BESA Research 6.1
- 3.1 MRI Segmentation
- 3.2 Load Sample EEG Data in BESA Research
- 3.3 Coregistration of EEG and MRI Data
- 3.4 Create new Folder for Custom Template Head-Model
- 3.5 Copy Files to Custom Template Head-Model Folder
- 3.6 Save Electrode Coordinates File
- 3.7 Create Talairach Transformation File
- 3.8 Rename Template Head-Model Files
- 3.9 Edit template_head_models.ini
- 3.10 Use your custom Template Head-Model in BESA Research 6.1
Introduction
This tutorial will guide you through the steps of how to create a custom template head-model in order to use it in the Source Analysis module of BESA Research 6.1.
Prerequisites
To create the template, you need to have the latest versions of BESA Research 6.1 and BESA MRI 2.0 installed on your computer. MRI data needs to be prepared to be available in either DICOM, Analyze or Nifti format.
The segmentation is a fully automatic, robust procedure which in general delivers very accurate segmentations. In some rare cases, however, it might happen that the automatic segmentation result contains inaccuracies and might, thus, be considered as inadequate. Segmentation problems might occur for data sets with atypical anatomy, for example, data sets containing brain lesions, as well as infant data sets. An alternative way of generating an adequate segmentation is to manually correct the segmentation mask, and then use the corrected segmentation mask as the basis for the FE mesh generation. This approach is well described on the following BESA Wiki website: Correcting Volume Conductor Segmentations
Custom Template Head-Model Integration in BESA Research 6.1
MRI Segmentation
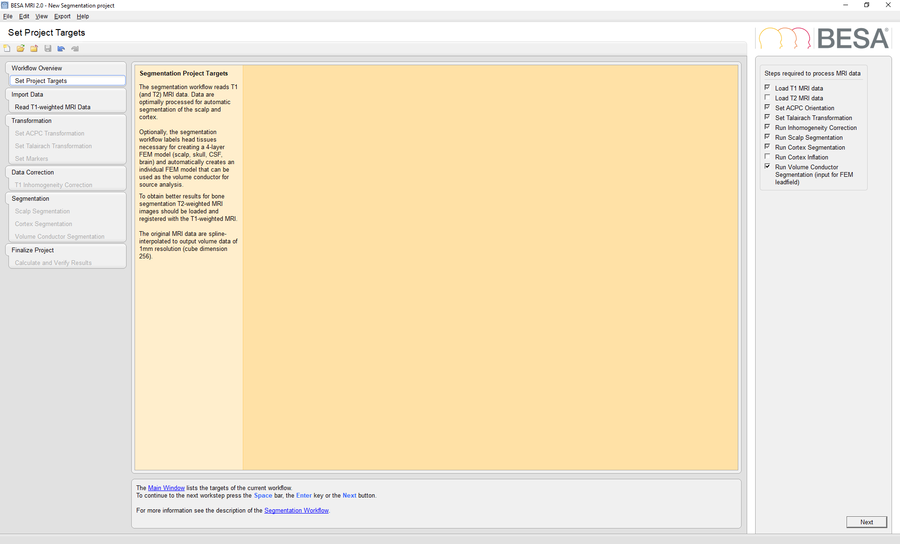
Start BESA MRI 2.0 and run a new segmentation project to perform a segmentation of the template MRI data in BESA MRI. Please mind that is necessary to check the option Run Volume Conductor Segmentation in the Set Project Targets workstep.
Load Sample EEG Data in BESA Research
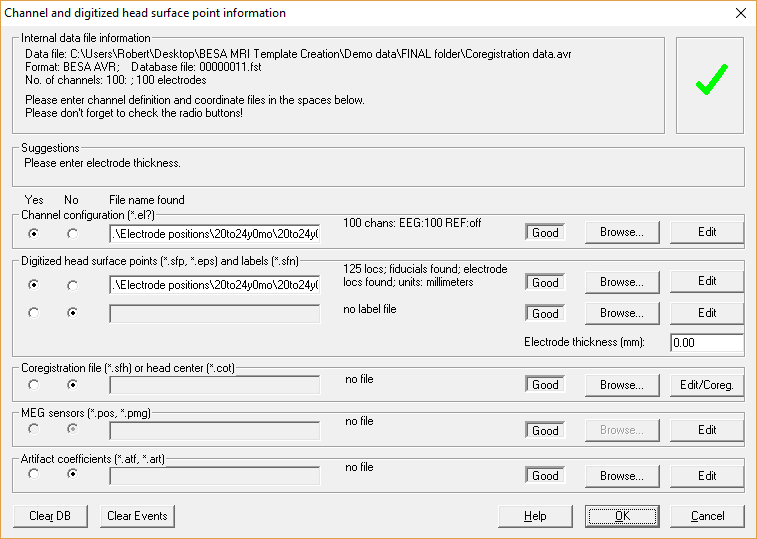
Open BESA Research and load the file Coregistration data.avr. To add electrode coordinates, navigate to File -> Head Surface Points and Sensors -> Load Coordinate Files.... Press Browse in the Channel configuration (*.el?) section and select the age-appropriate *.elp file from the Electrode positions for coregistration folder. Also load the age-appropriate *.sfp file by pressing Browse in the Digitized head surface points (*.sfp, *.eps) and labels (*.sfn) section and select the respective *.sfp file.
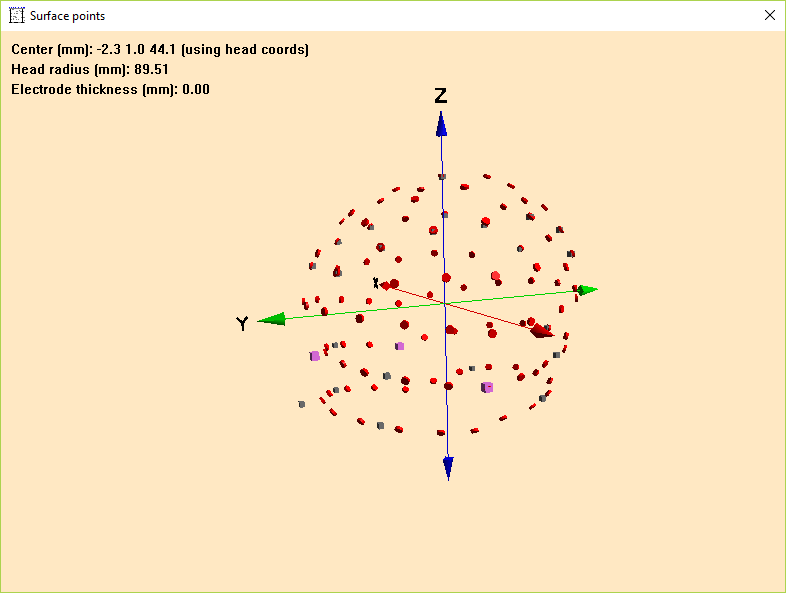
Please note that the coordinates of the electrode configuration will be fitted during coregistration and need not to have a perfect match with your template head-model at this step. Confirm all settings and leave this dialog via pressing OK. To check if all files were read correctly, open the Surface Points dialog (press V on your keyboard or select File -> Head Surface Points and Sensors -> View).
Coregistration of EEG and MRI Data
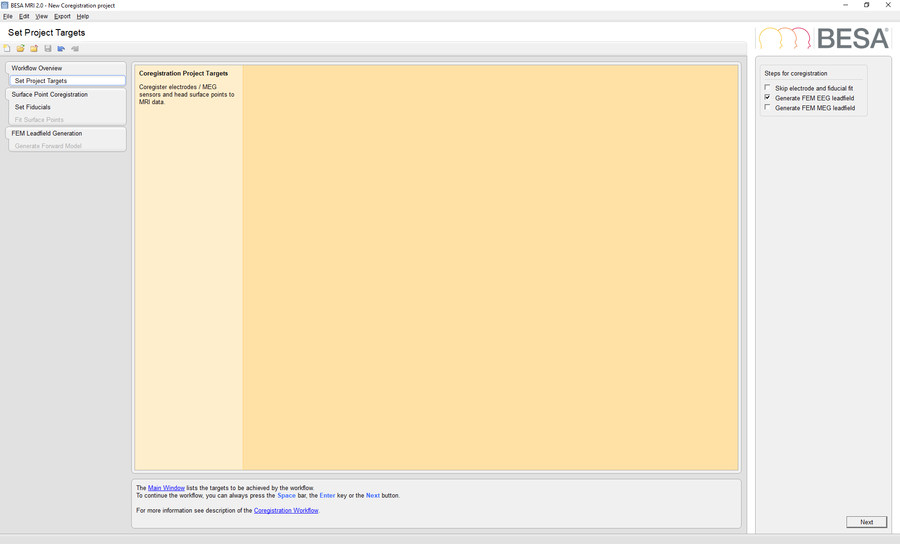
For coregistration of the sample EEG data set with your template head-model, select File -> Coregistration. Store the *.sfh file with the proposed filename. BESA MRI will now be started and a new coregistration project is preselected. If it is not started, press the Start New Coregistration button. Check the option Generate FEM EEG Leadfield in the Set Project Targets workstep and select the previously segmented MRI project as input for coregistration. Load the electrode configuration file Coregistration data.sfh in the Fit Surface Points workstep. Finalize the coregistration project by choosing the desired conductivity values, run the FEM computation and finally save the finished project.
Create new Folder for Custom Template Head-Model
To add a new template head-model for BESA Research, navigate to the installation directory. The default installation directory for BESA Research 6.1 is:
C:\Program Files (x86)\BESA\Research_6_1\
and create a new folder (e.g. 0y3mo) in the .\System\TemplateHeadModels\AgeAppropriateTemplates\ subfolder.