Difference between revisions of "Export"
(→Export Dialog) |
|||
| (11 intermediate revisions by 2 users not shown) | |||
| Line 2: | Line 2: | ||
|title = Module information | |title = Module information | ||
|module = BESA Research Basic or higher | |module = BESA Research Basic or higher | ||
| − | |version = 6.1 or higher | + | |version = BESA Research 6.1 or higher |
}} | }} | ||
| Line 12: | Line 12: | ||
* ASCII vectorized (short files only) | * ASCII vectorized (short files only) | ||
* EDF+ | * EDF+ | ||
| − | * | + | * Simple floating point matrix (e.g. for exporting data to Matlab) |
* Send the data directly to Matlab | * Send the data directly to Matlab | ||
| Line 19: | Line 19: | ||
* The entire data set or between markers | * The entire data set or between markers | ||
* The currently marked segment | * The currently marked segment | ||
| − | * Epochs around triggers | + | * [[Export Single Trial Data|Epochs around triggers]] |
* Standard deviations from binary average files (<span style="color:#ff9c00;">'''*.fsg'''</span>) | * Standard deviations from binary average files (<span style="color:#ff9c00;">'''*.fsg'''</span>) | ||
| Line 31: | Line 31: | ||
On export, you can choose to change the sampling rate. | On export, you can choose to change the sampling rate. | ||
| − | If exporting to BESA's binary format, you can optionally append the data to a preexisting data set, if sampling rates and the number of channels match | + | If exporting to BESA's binary format, you can optionally append the data to a preexisting data set, if sampling rates and the number of channels match. |
| − | + | To start export you have to: | |
* select File / Export... | * select File / Export... | ||
* press the <span style="color:#3366ff;">'''WrS '''</span>button. | * press the <span style="color:#3366ff;">'''WrS '''</span>button. | ||
| − | |||
* for Send to Matlab only: select File / Send To MATLAB... This also opens the Export Dialog, but only the <span style="color:#3366ff;">'''Send To Matlab radio'''</span> button is enabled as target format in the dialog. | * for Send to Matlab only: select File / Send To MATLAB... This also opens the Export Dialog, but only the <span style="color:#3366ff;">'''Send To Matlab radio'''</span> button is enabled as target format in the dialog. | ||
| Line 49: | Line 48: | ||
The dialog is started when you select'' File / Export...'' or press the <span style="color:#3366ff;">'''WrS'''</span> button or select '''Write Segment''' in the right click context menu when a segment has been highlighted. | The dialog is started when you select'' File / Export...'' or press the <span style="color:#3366ff;">'''WrS'''</span> button or select '''Write Segment''' in the right click context menu when a segment has been highlighted. | ||
| − | The dialog is divided into four sections | + | The dialog is divided into four sections: |
* ''Data to export''. Describes which data are to be exported. | * ''Data to export''. Describes which data are to be exported. | ||
* ''Montage and Filters.'' Which montage is to be exported, and whether or not filters are used. | * ''Montage and Filters.'' Which montage is to be exported, and whether or not filters are used. | ||
* ''Target data formats''. Specify the format of the exported data. | * ''Target data formats''. Specify the format of the exported data. | ||
* ''Resampling.'' Specify a new sampling rate in the exported data. | * ''Resampling.'' Specify a new sampling rate in the exported data. | ||
| + | |||
| + | ''Anonymize using target file name'': Export can be performed either with or without anonymizing using the checkbox "Anonymize using target file name". When the checkbox is checked, the subject name is replaced by a target file name, and the subject birth date is replaced by 1st July of the subject birth year. | ||
== Type of data to export == | == Type of data to export == | ||
| Line 64: | Line 65: | ||
* '''Continuous data'''. The whole data set or the data between markers are exported. | * '''Continuous data'''. The whole data set or the data between markers are exported. | ||
* '''Marked segment.''' If a segment of data is highlighted, this radio button is enabled. Select Marked segment to export just this segment. | * '''Marked segment.''' If a segment of data is highlighted, this radio button is enabled. Select Marked segment to export just this segment. | ||
| − | * '''Epochs around triggers.''' Export data segments around triggers. If this item is selected, the two buttons <span style="color:#3366ff;">'''Interval'''</span>... and <span style="color:#3366ff;">'''Triggers'''</span>... are enabled. They allow to select the interval (cf ''Edit / Default'' ''Block Epoch''...), and choose among the available triggers (see ''Edit / Trigger Values''...). Note that the default block epoch values are persistent across BESA Research sessions. The trigger selection is not persistent! | + | * '''Epochs around triggers.''' Export data segments around triggers. If this item is selected, the two buttons <span style="color:#3366ff;">'''Interval'''</span>... and <span style="color:#3366ff;">'''Triggers'''</span>... are enabled. They allow to select the interval (cf ''Edit / Default'' ''Block Epoch''...), and choose among the available triggers (see ''Edit / Trigger Values''...). Note that the default block epoch values are persistent across BESA Research sessions. The trigger selection is not persistent! You can read more about this option here: [[Export Single Trial Data]] |
* '''Standard Deviations (from fsg file only).''' If the average file was generated using the BESA Research ERP module, standard deviations are saved in the file. Check this item to export these values to an ASCII file. | * '''Standard Deviations (from fsg file only).''' If the average file was generated using the BESA Research ERP module, standard deviations are saved in the file. Check this item to export these values to an ASCII file. | ||
| Line 90: | Line 91: | ||
* '''Standard 81 electrode locations.''' EEG data are interpolated to a set of 81 electrodes on a standard head (average over 24 mainly Caucasian heads). | * '''Standard 81 electrode locations.''' EEG data are interpolated to a set of 81 electrodes on a standard head (average over 24 mainly Caucasian heads). | ||
| − | * Export can be performed either with or without the currently selected filters. Press the <span style="color:#3366ff;">'''Filters'''</span>... button to change the current filter settings (see also ''Filters / Edit Filter Settings''...). | + | * Export can be performed either with or without the currently selected filters. Press the <span style="color:#3366ff;">'''Filters'''</span>... button to change the current filter settings (see also ''Filters / Edit Filter Settings''...). Note that if artifact correction was performed and this checkbox is enabled, data will be exported with artifact correction. |
== Target Data Formats == | == Target Data Formats == | ||
| Line 100: | Line 101: | ||
The following target formats are available: | The following target formats are available: | ||
| − | * '''BESA binary''': Data are saved with the extension "<span style="color:#ff9c00;">'''.foc'''</span>" or "<span style="color:#ff9c00;">'''.fsg'''</span>". You can select whether to export with no compression (recommended for averages), or compressed (recommended for raw data). Note that compression can result in loss of resolution in averaged data. See | + | * '''BESA binary''': Data are saved with the extension "<span style="color:#ff9c00;">'''.foc'''</span>" or "<span style="color:#ff9c00;">'''.fsg'''</span>". You can select whether to export with no compression (recommended for averages), or compressed (recommended for raw data). Note that compression can result in loss of resolution in averaged data. See [[Export#Data_Compression|Data Compression]]. |
| − | * '''ASCII multiplexed''': Data are normally saved as text with the extension "<span style="color:#ff9c00;">'''.mul'''</span>". See | + | * '''ASCII multiplexed''': Data are normally saved as text with the extension "<span style="color:#ff9c00;">'''.mul'''</span>". See [[ASCII_File_Format#ASCII_Multiplexed_file_format|ASCII multiplexed format]] for a description of the data format. If the type of data to export is '''Epochs''' '''around triggers''', one file is exported for each trigger, and the file extension is a number, starting with ".000", and continuing ".001", ".002", etc. |
| − | * '''ASCII vectorized''': Data are normally saved as text with the extension "<span style="color:#ff9c00;">'''.avr'''</span>". See | + | * '''ASCII vectorized''': Data are normally saved as text with the extension "<span style="color:#ff9c00;">'''.avr'''</span>". See [[ASCII_File_Format#ASCII_Vectorized_file_format|ASCII vectorized format]] for a description of the data format. If the type of data to export is '''Epochs around triggers''', one file is exported for each trigger, and the file extension is a number, starting with ".000", and continuing ".001", ".002", etc. The data are not average referenced before saving. They will only be average referenced if the data are exported using an average referenced '''Current montage'''. You are only allowed to export small segments in vectorized format. If you have selected '''Continuous data''', this item will be disabled if the data are longer than 20 s in duration. |
* '''European Data Format (EDF+)''': Data are saved with the extension "<span style="color:#ff9c00;">'''.edf'''</span>". Export of '''Epochs around triggers''' to EDF+ is currently not possible. | * '''European Data Format (EDF+)''': Data are saved with the extension "<span style="color:#ff9c00;">'''.edf'''</span>". Export of '''Epochs around triggers''' to EDF+ is currently not possible. | ||
| − | * '''Simple binary matrix''': Data are written to a floating point binary matrix with the extension "<span style="color:#ff9c00;">'''.dat'''</span>". The matrix has the dimension no of samples x no of channels. In addition, a header file with the extension "<span style="color:#ff9c00;">'''.generic'''</span>" is also written. The header file is a text file contains information about the number of channels, sampling rate, and number of samples. These follow the specifications of the Generic File Format | + | * '''Simple binary matrix''': Data are written to a floating point binary matrix with the extension "<span style="color:#ff9c00;">'''.dat'''</span>". The matrix has the dimension no of samples x no of channels. In addition, a header file with the extension "<span style="color:#ff9c00;">'''.generic'''</span>" is also written. The header file is a text file contains information about the number of channels, sampling rate, and number of samples. These follow the specifications of the Generic File Format. This data format can be useful as a means of transferring data to other programs (e.g. '''MATLAB''') in a relatively compact form. If the type of data to export is '''Epochs around triggers''', the epochs are concatenated in the same target file. In this case, the header file contains a line specifying the number of epochs (cf. ''Generic File Format''). The number of samples in each epoch is the total number of samples, divided by the number of epochs. |
* '''Send To MATLAB''': The data are exported directly to MATLAB into the struct variable <code>besa_channels</code>. For more information on the data transfer from BESA Research to MATLAB, please refer to the Help chapter [[MATLAB_Interface | MATLAB interface]]. | * '''Send To MATLAB''': The data are exported directly to MATLAB into the struct variable <code>besa_channels</code>. For more information on the data transfer from BESA Research to MATLAB, please refer to the Help chapter [[MATLAB_Interface | MATLAB interface]]. | ||
| Line 137: | Line 138: | ||
== Data Compression == | == Data Compression == | ||
| − | When exporting to BESA binary format, you can compress the data to save space. Here we describe properties and pitfalls of the compression algorithm. | + | When exporting to BESA binary format and ASCII multiplex formats, you can compress the data to save space. Here we describe properties and pitfalls of the compression algorithm. |
| + | === Compression of BESA binary data === | ||
'''How compression works''' | '''How compression works''' | ||
| Line 154: | Line 156: | ||
For raw EEG data, we recommend using a step size of 0.5 µV. | For raw EEG data, we recommend using a step size of 0.5 µV. | ||
| − | |||
'''Different compression parameters for different data types (pitfalls!)''' | '''Different compression parameters for different data types (pitfalls!)''' | ||
| Line 165: | Line 166: | ||
* '''Averages:''' We recommend that averaged ERPs are not compressed. Since the signals are generally much smaller than the raw data, compression will lead to unacceptable loss of data resolution. | * '''Averages:''' We recommend that averaged ERPs are not compressed. Since the signals are generally much smaller than the raw data, compression will lead to unacceptable loss of data resolution. | ||
| + | |||
| + | === Compression of ASCII multiplexed data === | ||
| + | |||
| + | When compression is specified, data are written as integer values, and the scaling is specified in the header line of the *.mul file as ''Bins/uV=xxx''. | ||
| + | |||
| + | With no compression, the header contains ''Bins/uV=1.0'', and the data are written with 7 characters per number (e.g. -4.5084, 2.57239). | ||
| + | |||
| + | With compression, the data are written as integers, e.g. 0 -10 -6 -4, which can lead to considerably less characters and thus | ||
| + | |||
| + | {| class="wikitable" style="text-align:center;" | ||
| + | |- | ||
| + | ! No compression | ||
| + | ! 0.1 | ||
| + | ! 0.2 | ||
| + | ! 0.5 | ||
| + | ! 1.0 | ||
| + | |- | ||
| + | | -4.5084 | ||
| + | | -4.5 (-45) | ||
| + | | -4.6 (-23) | ||
| + | | -4.5 (-9) | ||
| + | | -5.0 (-5) | ||
| + | |- | ||
| + | | 2.57239 | ||
| + | | 2.6 (26) | ||
| + | | 2.6 (13) | ||
| + | | 2.5 (5) | ||
| + | | 3.0 (3) | ||
| + | |} | ||
| + | |||
| + | The numbers in brackets are the values written to the data file. | ||
[[Category:Research Manual]] | [[Category:Research Manual]] | ||
{{BESAManualNav}} | {{BESAManualNav}} | ||
Latest revision as of 15:59, 5 May 2021
| Module information | |
| Modules | BESA Research Basic or higher |
| Version | BESA Research 6.1 or higher |
Contents
Exporting files from BESA
Data can be exported from BESA Research in the following formats:
- BESA's own binary format (compressed or uncompressed)
- ASCII multiplexed
- ASCII vectorized (short files only)
- EDF+
- Simple floating point matrix (e.g. for exporting data to Matlab)
- Send the data directly to Matlab
You can export
- The entire data set or between markers
- The currently marked segment
- Epochs around triggers
- Standard deviations from binary average files (*.fsg)
Export is allowed to various montages:
- filtered or unfiltered data
- original data
- current montage (including artifact correction if applied)
- standard 81-electrode montage.
On export, you can choose to change the sampling rate.
If exporting to BESA's binary format, you can optionally append the data to a preexisting data set, if sampling rates and the number of channels match.
To start export you have to:
- select File / Export...
- press the WrS button.
- for Send to Matlab only: select File / Send To MATLAB... This also opens the Export Dialog, but only the Send To Matlab radio button is enabled as target format in the dialog.
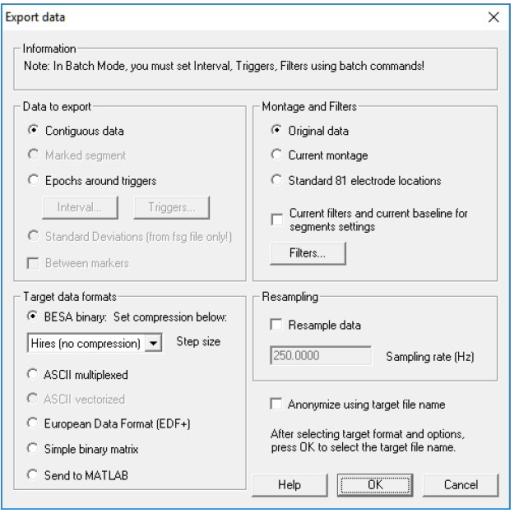
Export Dialog
The dialog is started when you select File / Export... or press the WrS button or select Write Segment in the right click context menu when a segment has been highlighted.
The dialog is divided into four sections:
- Data to export. Describes which data are to be exported.
- Montage and Filters. Which montage is to be exported, and whether or not filters are used.
- Target data formats. Specify the format of the exported data.
- Resampling. Specify a new sampling rate in the exported data.
Anonymize using target file name: Export can be performed either with or without anonymizing using the checkbox "Anonymize using target file name". When the checkbox is checked, the subject name is replaced by a target file name, and the subject birth date is replaced by 1st July of the subject birth year.
Type of data to export
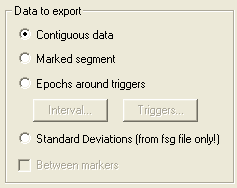
The types of data to export are:
- Continuous data. The whole data set or the data between markers are exported.
- Marked segment. If a segment of data is highlighted, this radio button is enabled. Select Marked segment to export just this segment.
- Epochs around triggers. Export data segments around triggers. If this item is selected, the two buttons Interval... and Triggers... are enabled. They allow to select the interval (cf Edit / Default Block Epoch...), and choose among the available triggers (see Edit / Trigger Values...). Note that the default block epoch values are persistent across BESA Research sessions. The trigger selection is not persistent! You can read more about this option here: Export Single Trial Data
- Standard Deviations (from fsg file only). If the average file was generated using the BESA Research ERP module, standard deviations are saved in the file. Check this item to export these values to an ASCII file.
Between markers. If there are markers in the file, and Marked segment is not selected, selecting Between markers will result in the export of data between markers relative to the current position in the file (as defined by the middle of the current display), either:
- if there is no previous marker, from the beginning of the file to the next marker, or
- from the previous to the next marker, or
- if there is no next marker, from the previous marker to the end of the file.
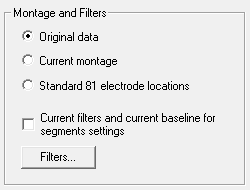
Montages and Filters
The data can be exported either
- Original data. The data are exported using the original montage
- Current montage. The currently selected montage is exported. If extra channels, e.g. selected channels or artifact waveforms are displayed, these are exported as well. Notes:
- When 'Current Montage' is selected, no auxiliary files are exported. When re-importing the data into BESA Research, all channels will be defined as polygraphic.
- If the current data is artifact-corrected, the artifact-corrected data will be exported when 'Current Montage' is selected.
- Standard 81 electrode locations. EEG data are interpolated to a set of 81 electrodes on a standard head (average over 24 mainly Caucasian heads).
- Export can be performed either with or without the currently selected filters. Press the Filters... button to change the current filter settings (see also Filters / Edit Filter Settings...). Note that if artifact correction was performed and this checkbox is enabled, data will be exported with artifact correction.
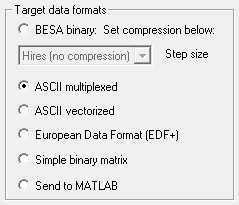
Target Data Formats
The following target formats are available:
- BESA binary: Data are saved with the extension ".foc" or ".fsg". You can select whether to export with no compression (recommended for averages), or compressed (recommended for raw data). Note that compression can result in loss of resolution in averaged data. See Data Compression.
- ASCII multiplexed: Data are normally saved as text with the extension ".mul". See ASCII multiplexed format for a description of the data format. If the type of data to export is Epochs around triggers, one file is exported for each trigger, and the file extension is a number, starting with ".000", and continuing ".001", ".002", etc.
- ASCII vectorized: Data are normally saved as text with the extension ".avr". See ASCII vectorized format for a description of the data format. If the type of data to export is Epochs around triggers, one file is exported for each trigger, and the file extension is a number, starting with ".000", and continuing ".001", ".002", etc. The data are not average referenced before saving. They will only be average referenced if the data are exported using an average referenced Current montage. You are only allowed to export small segments in vectorized format. If you have selected Continuous data, this item will be disabled if the data are longer than 20 s in duration.
- European Data Format (EDF+): Data are saved with the extension ".edf". Export of Epochs around triggers to EDF+ is currently not possible.
- Simple binary matrix: Data are written to a floating point binary matrix with the extension ".dat". The matrix has the dimension no of samples x no of channels. In addition, a header file with the extension ".generic" is also written. The header file is a text file contains information about the number of channels, sampling rate, and number of samples. These follow the specifications of the Generic File Format. This data format can be useful as a means of transferring data to other programs (e.g. MATLAB) in a relatively compact form. If the type of data to export is Epochs around triggers, the epochs are concatenated in the same target file. In this case, the header file contains a line specifying the number of epochs (cf. Generic File Format). The number of samples in each epoch is the total number of samples, divided by the number of epochs.
- Send To MATLAB: The data are exported directly to MATLAB into the struct variable
besa_channels. For more information on the data transfer from BESA Research to MATLAB, please refer to the Help chapter MATLAB interface.
Resampling
Check Resample data to change the sampling rate of the target data. The edit box is enabled, and you specify a sampling rate.
Data are resampled using splines. Thus, the new sampling rate is not limited to fractions or multiples of the original rate.
Note that if Resample data is not checked, the sampling rate of the source data is displayed.
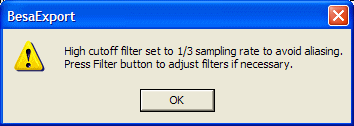
Resampling and aliasing. If you want to reduce the sampling rate it is important to avoid aliasing! It is recommended that a low-pass filter with a boundary frequency of not more than 1/3 of the original sampling rate is used. When you set a new sampling rate, BESA Research checks the current filter settings. If export without filters is selected, or if the current low-pass filter is set to a value that is higher than 1/3 of the sampling rate, BESA Research sets the filter, and opens a message box with a warning:
You may adjust the filters by pressing the Filter button, e.g. if the original data were already recorded with a sufficiently low low-pass filter setting, so that additional filtering is unnecessary.
Data Compression
When exporting to BESA binary format and ASCII multiplex formats, you can compress the data to save space. Here we describe properties and pitfalls of the compression algorithm.
Compression of BESA binary data
How compression works
The compression algorithm works on two principals:
- Data resolution can often be reduced without losing data quality. For instance, a data resolution of 0.1 µV or higher is unnecessary for viewing normal EEG -- 0.5 µV or 1 µV steps are sufficient. Similarly for event-related potentials: the raw data only require a resolution of 0.5 µV or 1 µV to achieve a much higher resolution after averaging.
- Differences between successive data samples on a signal are generally much smaller than the absolute values of the data. Thus, one start value, and then a series of subsequent differences can be stored in a much smaller space than an equivalent series of absolute values. A consequence of this principle is that smoothed signals (with high frequencies removed) can be compressed into a smaller space than signals with a lot of high frequency noise.
Compression parameter
The parameter you need to choose is the data resolution, or step size. Steps smaller than this size will no longer be represented in the compressed data. The BESA Research Export Module allows the following steps for the compression of EEG signals:
0.1 µV, 0.2 µV, 0.5 µV, 1 µV
For raw EEG data, we recommend using a step size of 0.5 µV.
Different compression parameters for different data types (pitfalls!)
As described above, the step sizes make sense for EEG signals. For other types of data, other step sizes make more sense. In addition, a polygraphic signal can have the same order of signal magnitude as the EEG (e.g. an EOG or EKG signal), but it might have a completely different scale, e.g. a voice signal, recorded in mV or V. To help accommodate different orders of signal magnitude, BESA Research applies the following rules:
- MEG data: Step sizes (in units of fT) are 20 x the step size in µV. Thus, a step size of 0.5 µV will lead to a step size of 10 fT for MEG data.
- Polygraphic and ICR data: The step size depends on the current amplitude scaling factor in BESA Research. A multiplication factor is used that is the current scaling factor, divided by 100. Thus, if the scale is set to 1 V, the factor is 10 mV. If you have chosen an EEG step size of 0.5 µV, the resulting step size will be 10 x 0.5 = 5 mV.
A pitfall in compression is that if the current amplitude scaling for polygraphic or ICR data does not display the signal sensibly, compression may lead to complete loss of the signal. Note that this only applies to polygraphic and ICR channel types.
- Averages: We recommend that averaged ERPs are not compressed. Since the signals are generally much smaller than the raw data, compression will lead to unacceptable loss of data resolution.
Compression of ASCII multiplexed data
When compression is specified, data are written as integer values, and the scaling is specified in the header line of the *.mul file as Bins/uV=xxx.
With no compression, the header contains Bins/uV=1.0, and the data are written with 7 characters per number (e.g. -4.5084, 2.57239).
With compression, the data are written as integers, e.g. 0 -10 -6 -4, which can lead to considerably less characters and thus
| No compression | 0.1 | 0.2 | 0.5 | 1.0 |
|---|---|---|---|---|
| -4.5084 | -4.5 (-45) | -4.6 (-23) | -4.5 (-9) | -5.0 (-5) |
| 2.57239 | 2.6 (26) | 2.6 (13) | 2.5 (5) | 3.0 (3) |
The numbers in brackets are the values written to the data file.
| Review | |
|---|---|
| Source Analysis | |
| Integration with MRI and fMRI | |
| Source Coherence | |
| Export |
|
| MATLAB Interface | |
| Special Topics | |