Difference between revisions of "The Initialization File: BESA.ini"
m |
m |
||
| (24 intermediate revisions by 4 users not shown) | |||
| Line 2: | Line 2: | ||
|title = Module information | |title = Module information | ||
|module = BESA Research Basic or higher | |module = BESA Research Basic or higher | ||
| − | |version = 6.1 or higher | + | |version = BESA Research 6.1 or higher |
}} | }} | ||
| Line 9: | Line 9: | ||
'''BESA.ini File''' | '''BESA.ini File''' | ||
| − | BESA Research uses settings provided in the initialization file <span style="color:#ff9c00;">'''BESA.ini'''</span> whenever BESA Research is started or a new file is opened for the first time. The format of this file conforms with standard initialization files (<span style="color:#ff9c00;">'''<nowiki>*.ini</nowiki>'''</span>) of Windows. You may change the settings in <span style="color:#ff9c00;">'''BESA.ini'''</span> using <span style="color:#ff9c00;"><span style="color:#00000a;"> | + | BESA Research uses settings provided in the initialization file <span style="color:#ff9c00;">'''BESA.ini'''</span> whenever BESA Research is started or a new file is opened for the first time. The format of this file conforms with standard initialization files (<span style="color:#ff9c00;">'''<nowiki>*.ini</nowiki>'''</span>) of Windows. You may change the settings in <span style="color:#ff9c00;">'''BESA.ini'''</span> using <span style="color:#ff9c00;"><span style="color:#00000a;">Notepad.exe</span></span> from the ACCESSORIES group, or other plain text editors to adapt BESA Research to '''your own everyday needs'''. The default settings provided in <span style="color:#ff9c00;">'''BESA.ini'''</span> will be used by BESA Research whenever BESA Research or the launch program is started. It is advised that you make a backup copy of <span style="color:#ff9c00;">'''BESA.ini'''</span> before you change the default settings. |
| Line 16: | Line 16: | ||
You can place <span style="color:#ff9c00;">'''BESA.ini'''</span> at three possible locations: | You can place <span style="color:#ff9c00;">'''BESA.ini'''</span> at three possible locations: | ||
| − | # '''Private''': each user on a PC should have his/her own private settings. This is normally in '' | + | # '''Private''': each user on a PC should have his/her own private settings. This is normally in ''Documents/BESA/Research_7_1'' |
| − | # '''Public''': all users should use one setting, but they can edit <span style="color:#ff9c00;">'''BESA.ini'''</span> to change the settings. This is normally in ''Public Documents/BESA/ | + | # '''Public''': all users should use one setting, but they can edit <span style="color:#ff9c00;">'''BESA.ini'''</span> to change the settings. This is normally in ''Public Documents/BESA/Research_7_1'' |
| − | # '''Administrator''': the PC administrator determines the settings. This is normally in ''C:Program Files(x86)/BESA/ | + | # '''Administrator''': the PC administrator determines the settings. This is normally in ''C:Program Files(x86)/BESA/Research_7_1'' |
The actual folder names depend on the operating system and the system language. | The actual folder names depend on the operating system and the system language. | ||
| Line 27: | Line 27: | ||
'''There are 13 general sections, and several reader-specific sections:''' | '''There are 13 general sections, and several reader-specific sections:''' | ||
| − | [Defaults] | + | {| class="wikitable" |
| − | + | | [Defaults] | |
| − | [Folders] | + | | General settings (filters, scaling, and various other settings) |
| − | + | |- | |
| − | [Electrodes] | + | | [Folders] |
| − | + | | Folders used by BESA Research (Examples, Montages, Scripts, Settings,...) | |
| − | [Patterns] | + | |- |
| − | + | | [Electrodes] | |
| − | [Artifacts] | + | | Electrode renaming |
| − | + | |- | |
| − | [KEYCONTROLS] | + | | [Patterns] |
| − | + | | Rename patterns in the <span style="color:#3366ff;">'''Tags'''</span> menu | |
| − | [Search] | + | |- |
| − | + | | [Artifacts] | |
| − | [FFT] | + | | Settings for artifact correction |
| − | + | |- | |
| − | [Printer] | + | | [KEYCONTROLS] |
| − | + | | Function key definitions | |
| − | [Calibration] | + | |- |
| − | + | | [Search] | |
| − | [Video] | + | | Default parameters for search |
| − | + | |- | |
| − | [Mapping] | + | | [FFT] |
| − | + | | Frequency band definitions | |
| − | [Matlab] | + | |- |
| − | + | | [Printer] | |
| − | [ | + | | Printer control |
| − | + | |- | |
| + | | [Calibration] | ||
| + | | Calibration control | ||
| + | |- | ||
| + | | [Video] | ||
| + | | Digital video control | ||
| + | |- | ||
| + | | [Mapping] | ||
| + | | Mapping control | ||
| + | |- | ||
| + | | [Updates] | ||
| + | | Options for program updates | ||
| + | |- | ||
| + | | [Matlab] | ||
| + | | Settings for the MATLAB interface | ||
| + | |- | ||
| + | | [fMRI] | ||
| + | | Settings for the fMRI arfifact removal | ||
| + | |- | ||
| + | | [Montages] | ||
| + | | A setting for a default source montage | ||
| + | |} | ||
'''Reader-specific settings''' | '''Reader-specific settings''' | ||
| Line 75: | Line 96: | ||
[XLTEK] | [XLTEK] | ||
| − | |||
== Defaults == | == Defaults == | ||
| − | + | <br> | |
'''Default settings provided for section [Defaults]:''' | '''Default settings provided for section [Defaults]:''' | ||
| Line 86: | Line 106: | ||
'''DataBuffering=Off''' (If set to "On", an internal buffer of length 180 s of data is kept to speed up paging). This can speed up paging, particularly when the data are in a network folder. | '''DataBuffering=Off''' (If set to "On", an internal buffer of length 180 s of data is kept to speed up paging). This can speed up paging, particularly when the data are in a network folder. | ||
| − | '''DisplayedTime=10''' | + | '''DisplayedTime=10''' displayed time window [s] on the screen |
| − | '''Montage=Org''' | + | '''Montage=Org''' montage used when opening a new file |
| − | '''ScpScale=50''' | + | '''ScpScale=50''' scale of scalp channels in [mV] |
| − | '''PgrScale=500''' | + | '''PgrScale=500''' scale of polygraphic channels in [mV] |
| − | '''IcrScale=500''' | + | '''IcrScale=500''' scale of intracranial channels in [mV] |
| − | '''MegScale= | + | '''MegScale=200''' scale of MEG/GRA channels in [fT or fT/cm] |
| − | ''' | + | '''MagScale=1000''' scale of MAG channels in [fT] (''this feature requires BESA Research 7.1 or higher'') |
| − | ''' | + | '''SrcScale=100''' scale of source of source montages |
| − | ''' | + | '''BaselineCorrection=On''' baseline correction, do not switch off in AC systems |
| − | ''' | + | '''ClippingPercent= '''set from 100 to 200 if you want to clip artifacts in displayed EEG (not used if empty or 0) |
| − | ''' | + | '''LowFilter=''' low filter cutoff frequency [Hz] (variable filter) |
| − | ''' | + | '''TimeConstant=0.3''' time constant for low filter cutoff frequency [sec] (fixed forward filter, 0.3 sec is equivalent to 0.53 Hz) |
| − | ''' | + | '''HighFilter=70''' high filter cutoff frequency [Hz] (variable filter) |
| − | ''' | + | '''NotchFilter=50''' notch filter center frequency [Hz] |
| − | '''BandFilterStatus=Off''' | + | '''NotchFilterStatus=Off''' notch filter is off, set=On if you want to use as default |
| + | |||
| + | '''BandFilter=12''' band pass filter center frequency [Hz] | ||
| + | |||
| + | '''BandFilterStatus=Off''' band pass is off, set=On if you want to use as default | ||
'''AdditionalChannelFile=''' defines the full path and name of an additional channels montage file, e.g. <span style="color:#ff9c00;">'''C:\Program Files\BESA\Research_x\Montages\AdditionalChannels\EKG.sel'''</span> | '''AdditionalChannelFile=''' defines the full path and name of an additional channels montage file, e.g. <span style="color:#ff9c00;">'''C:\Program Files\BESA\Research_x\Montages\AdditionalChannels\EKG.sel'''</span> | ||
| − | '''ColoredWaveforms=On''' | + | '''ColoredWaveforms=On''' scalp waveforms are (not) colored according to region |
| + | |||
| + | '''WriteSegmentPath=''' defines default path for saving segments/averages. If blank, the path of the current data file is used. | ||
| + | |||
| + | '''ShowSubjectInfo=Off''' subject info will (not) be displayed. | ||
| − | ''' | + | '''ParallelComputing=On''' defines if parallel computing during extensive computation should be used or not (''this feature requires BESA Research 7.1 or higher'') |
| − | ''' | + | '''MapSmoothing=0''' set a non-zero value to specify a default map smoothing parameter (normally specified in ''Options/Mapping/Spline Interpolation Smoothing Constant''). Valid values are within the range between 1e-8 and 1e-4. Values outside this range will be set to within the range. |
The following optional parameters are not defined as default and can be set manually in<span style="color:#ff9c00;">''' BESA.ini'''</span>: | The following optional parameters are not defined as default and can be set manually in<span style="color:#ff9c00;">''' BESA.ini'''</span>: | ||
| − | + | '''TextEditor="Notepad.exe"''' defines the path to your preferred text editor. This will be used when you press the <span style="color:#3366ff;">'''Edit'''</span> button in the ''Load Coordinate Files dialog box''. | |
| − | '''TextEditor="Notepad.exe"''' defines the path to your preferred text editor. This will be used when you press the <span style="color:#3366ff;">'''Edit'''</span> button the ''Load Coordinate Files dialog box''. | + | |
'''NeuroScanDataNumberOfBits=32''' defines the format of NeuroScan data files ('16' for 16-bit, '32' for 32-bit). If this variable is not specified, BESA uses a heuristic to (try to) decide which of the two data formats is used. This variable overrides the heuristic. If you want to specify the NeuroScan data format for specific files, create a file, named "16bit" or "32bit", and place it in the data folder. | '''NeuroScanDataNumberOfBits=32''' defines the format of NeuroScan data files ('16' for 16-bit, '32' for 32-bit). If this variable is not specified, BESA uses a heuristic to (try to) decide which of the two data formats is used. This variable overrides the heuristic. If you want to specify the NeuroScan data format for specific files, create a file, named "16bit" or "32bit", and place it in the data folder. | ||
| Line 144: | Line 171: | ||
If BaselineCorrection is set to 'On', before displaying a screen of data, BESA subtracts for each channel the mean over its displayed time points. This optimizes viewing, because it ensures that the vertical position of each channel is not shifted upward or downward from the channel label at the left of the screen. There are some cases in which you will not want baseline correction, i.e. when the DC level in the data is already correctly defined. This is usually the case, for instance, when reading in files that have been processed by BESA. In this case, BaselineCorrection should be set to 'Off', because otherwise maps and source montage displays may be distorted. | If BaselineCorrection is set to 'On', before displaying a screen of data, BESA subtracts for each channel the mean over its displayed time points. This optimizes viewing, because it ensures that the vertical position of each channel is not shifted upward or downward from the channel label at the left of the screen. There are some cases in which you will not want baseline correction, i.e. when the DC level in the data is already correctly defined. This is usually the case, for instance, when reading in files that have been processed by BESA. In this case, BaselineCorrection should be set to 'Off', because otherwise maps and source montage displays may be distorted. | ||
| − | |||
== Folders == | == Folders == | ||
| − | '''The [Folders] section defines where BESA Research places its files. In versions 5.1 and earlier, files were located in various subfolders of the program folder. This led to problems if the user did not have administrator rights, e.g. to create or write to a file | + | '''The [Folders] section defines where BESA Research places its files. In versions 5.1 and earlier, files were located in various subfolders of the program folder. This led to problems if the user did not have administrator rights, e.g. to create or write to a file. If you wish, you can also specify paths in the [Folders] section to use the previous locations. The previous location is given for each variable.''' |
These settings allow some flexibility that can be useful if you want to tune BESA Research for use by several users, or on a network. For instance, the Examples and Montages folders might be located on a network disk. For the current defaults, the database, Examples, Montages, and Scripts are set up for use by all users on the PC on which BESA Research is installed. The settings files (<span style="color:#ff9c00;">'''Besa.set'''</span>, <span style="color:#ff9c00;">'''Besa.cfg'''</span>, etc.) are located in private folders so that each user retains his or her own settings. | These settings allow some flexibility that can be useful if you want to tune BESA Research for use by several users, or on a network. For instance, the Examples and Montages folders might be located on a network disk. For the current defaults, the database, Examples, Montages, and Scripts are set up for use by all users on the PC on which BESA Research is installed. The settings files (<span style="color:#ff9c00;">'''Besa.set'''</span>, <span style="color:#ff9c00;">'''Besa.cfg'''</span>, etc.) are located in private folders so that each user retains his or her own settings. | ||
| Line 171: | Line 197: | ||
'''User=%privatprog%Settings''' The path for user defined settings (used to be ''%progdir%System\Userdirs'' in BESA versions up to 5.1.x) | '''User=%privatprog%Settings''' The path for user defined settings (used to be ''%progdir%System\Userdirs'' in BESA versions up to 5.1.x) | ||
| + | |||
| + | '''DataExport=%privateprog%Export''' The path for data to be exported for BESA Connectivity (not listed by default, but can be adjusted by the user) | ||
| Line 177: | Line 205: | ||
The strings enclosed by percent signs (%) are placeholders for the following folders in English-language versions of Windows. Folder names differ depending on Windows version, and for other language settings. BESA Research will substitute the placeholders by the appropriate folder name for the system and the system language: | The strings enclosed by percent signs (%) are placeholders for the following folders in English-language versions of Windows. Folder names differ depending on Windows version, and for other language settings. BESA Research will substitute the placeholders by the appropriate folder name for the system and the system language: | ||
| + | '''Windows 7, 8.1, and 10 (English):''' | ||
| − | + | '''%localapp%''' = "''C:\Users\[user]\Documents\BESA\Research_7_0''", where [user] is the logon name of the current user. This folder is directly accessible from the Desktop as "''Desktop\[user]\Documents\BESA\Research_7_0''". | |
| − | + | ||
| − | '''%localapp%''' = "''C:\Users\[user]\ | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | '''%publicprog%''' = "''C:\ | + | '''%publicprog%''' = "''C:\Users\Public\Public Documents\BESA\Research_7_0''". |
| − | '''%privateprog%''' = "''C:\ | + | '''%privateprog%''' = "''C:\Users\[user]\Documents\BESA\Research_7_0''", where [user] is the logon name of the current user. This folder is directly accessible from the Windows Explorer as "''Desktop\[User]\Documents\BESA\Research_7_0''". |
| − | '''%progdir%''' = the BESA Research root folder. In a default installation, this is "''C:\Program | + | '''%progdir%''' = the BESA Research root folder. In a default installation, this is "''C:\Program Files (x86)\BESA\Research_7_0''". |
'''%besaroot%''' is the same as '''%progdir%''' | '''%besaroot%''' is the same as '''%progdir%''' | ||
| Line 527: | Line 515: | ||
'''UseBitmapDrawing=Off''' | '''UseBitmapDrawing=Off''' | ||
| − | Set this to "On" if 3D maps show a strange pattern of black triangular shapes (this is frequently observed with modern Intel On-Board graphics controllers, and is a result of inadequate drivers for | + | Set this to "On" if 3D maps show a strange pattern of black triangular shapes (this is frequently observed with modern Intel On-Board graphics controllers, and is a result of inadequate drivers for OpenGL). |
'''Use3DVBlending=Auto''' | '''Use3DVBlending=Auto''' | ||
| Line 535: | Line 523: | ||
Set this to "On" if the 3D view in the Montage Editor or the Source Analysis window shows a ragged surface boundary. | Set this to "On" if the 3D view in the Montage Editor or the Source Analysis window shows a ragged surface boundary. | ||
| − | ''' | + | '''UseDoubleBuffering=On''' |
| − | Set | + | Set this to "Off" to disable double buffering mechanism that prevents the screen from flickering while paging through data and dragging window (''this feature requires BESA Research 7.1 or higher''). |
| + | |||
| + | Note: '''MapSmoothing''', the default map smoothing parameter, can be specified in the '''[Defaults]''' section. | ||
== Matlab == | == Matlab == | ||
| Line 545: | Line 535: | ||
'''Default settings for the [Matlab] section:''' | '''Default settings for the [Matlab] section:''' | ||
| − | ''' | + | '''Platform=64''' |
| − | + | ||
| − | + | ||
| + | Set '''Platform=32''' if you want to use the x86 version of MATLAB. | ||
== Updates == | == Updates == | ||
| Line 565: | Line 554: | ||
'''LocalPath''' | '''LocalPath''' | ||
| − | For the network administrator. This can be set to a path on the local network to the BESA update files, so that users can obtain their updates locally. The path is given to the text file "<span style="color:#ff9c00;">'''UpdateVersions.txt'''</span>" (e.g. ''LocalPath=\\transtec-sak\zarascratch\BESA\Updates\UpdateVersions.txt''), which contains further details for the program to obtain its updates. If you want to use this feature, please contact us | + | For the network administrator. This can be set to a path on the local network to the BESA update files, so that users can obtain their updates locally. The path is given to the text file "<span style="color:#ff9c00;">'''UpdateVersions.txt'''</span>" (e.g. ''LocalPath=\\transtec-sak\zarascratch\BESA\Updates\UpdateVersions.txt''), which contains further details for the program to obtain its updates. If you want to use this feature, please contact us using our [https://besagmbh.atlassian.net/servicedesk/customer/portals support portal]. |
The following variables are not required, because BESA Research has the paths hardwired: | The following variables are not required, because BESA Research has the paths hardwired: | ||
| Line 571: | Line 560: | ||
'''FTP1 (also FTP2, FTP3)''' | '''FTP1 (also FTP2, FTP3)''' | ||
| − | + | Download server | |
'''Path1 (also Path2, Path3)''' | '''Path1 (also Path2, Path3)''' | ||
| Line 586: | Line 575: | ||
| − | == | + | == FMRI == |
| + | ''(requires Besa Research 7.0 or higher)'' | ||
| − | ''' | + | These settings define the default parameters for the fMRI artifact removal in the BESA Research (see [[BESA_Research_Artifact_Correction#fMRI_artifact_removal|fMRI artifact removal]] chapter for further details). For example: |
| + | |||
| + | <syntaxhighlight lang="text"> | ||
| + | [FMRI] | ||
| + | FMRIRemovalMode=1 | ||
| + | TRDelay=200 | ||
| + | TRLength=800 | ||
| + | NumberOfAverages=21 | ||
| + | fMRImoveThreshold=0.15 | ||
| + | FMRITRID=8015 | ||
| + | ScansToSkip=0 | ||
| + | </syntaxhighlight> | ||
| + | |||
| + | These values indicate: | ||
| + | |||
| + | * '''FMRIRemovalMode''': Removal method (0: Turned off; 1: Allen et al, 2000; 2: Allen et al., 2000 Modified; 3: Moosmann et al.,2003) | ||
| + | * '''TRDelay''': Delay between marker and start of volume acquisition [ms] | ||
| + | * '''NumberOfAverages''': Number of artifact occurrence averages | ||
| + | * '''fMRImoveThreshold''': Movement threshold [mm] | ||
| + | * '''FMRITRID''': fMRI Trigger code | ||
| + | * '''ScansToSkip''': Number of scans to skip | ||
| + | |||
| + | |||
| + | == Montage == | ||
| + | |||
| + | '''The section [Montage] allows to specify an initial montage that is set the first time when the source (Src), recorded (Rec), virtual (Vir) or user (Usr) montage button is pressed. If BESA.ini does not specify a montage, pressing the corresponding button opens the drop-down menu offering all the available montages for the current montage type.''' | ||
| + | |||
| + | |||
| + | '''Source=25s''' specifies that when the Src button in the control ribbon is pressed for the first time, the source montage "25s" will be selected. | ||
| + | |||
| + | |||
| + | |||
| + | '''Recorded=Original Recording''' specifies that when the Rec button in the control ribbon is pressed for the first time, the source montage "Original Recording" will be selected. | ||
| + | |||
| + | |||
| + | |||
| + | '''Virtual=Triple Banana''' specifies that when the Vir button in the control ribbon is pressed for the first time, the source montage "Triple Banana" will be selected. | ||
| + | |||
| + | |||
| + | |||
| + | '''User=CA25''' specifies that when the Usr button in the control ribbon is pressed for the first time, the source montage "CA25" will be selected. | ||
| + | |||
| + | |||
| + | |||
| + | == Reader-Specific Settings == | ||
| + | <br> | ||
| + | === BrainLab === | ||
'''Default settings provided for section [BrainLab]:''' | '''Default settings provided for section [BrainLab]:''' | ||
| Line 597: | Line 633: | ||
| − | + | === Bio-Logic === | |
| − | + | ||
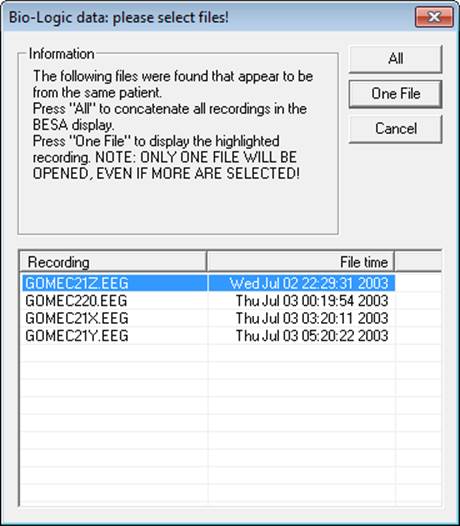
'''FileSelect=Yes''' | '''FileSelect=Yes''' | ||
If there are several Bio-Logic files in a data folder, the reader can check if the files have the same settings. There are three possible options: | If there are several Bio-Logic files in a data folder, the reader can check if the files have the same settings. There are three possible options: | ||
| + | |||
* Open a dialog to ask if the files should be treated as a single data set, or as individual, separate files. | * Open a dialog to ask if the files should be treated as a single data set, or as individual, separate files. | ||
| − | |||
[[Image:ST Besa ini (2).jpg ]] | [[Image:ST Besa ini (2).jpg ]] | ||
| + | <div style="margin-left:0.953cm;margin-right:0cm;">in this case, use '''FileSelect=Yes''' (this is the default setting) Note that the choice made in the dialog will apply to the file(s) within a BESA Research session. For a given file and session, the dialog will only be opened once, even if the file is closed and reopened.</div> | ||
| − | |||
* Always concatenate such files into a single data set. In this case use '''FileSelect=All''' | * Always concatenate such files into a single data set. In this case use '''FileSelect=All''' | ||
* Always open the files as single, separate files. In this case use '''FileSelect=Single''' | * Always open the files as single, separate files. In this case use '''FileSelect=Single''' | ||
| − | + | === EDF+/BDF/Trackit === | |
| − | + | ||
'''TriggerScan=On''' | '''TriggerScan=On''' | ||
| Line 622: | Line 656: | ||
| − | + | === EGI === | |
| − | + | ||
The treatment of DIN events can be modified in the''' [EGI] '''section: | The treatment of DIN events can be modified in the''' [EGI] '''section: | ||
| Line 633: | Line 666: | ||
'''SeparateDINevents'''<nowiki>=yes/no (default is “yes”)</nowiki> | '''SeparateDINevents'''<nowiki>=yes/no (default is “yes”)</nowiki> | ||
| − | Set to “no” if you don’t want to treat DIN events separately. | + | Set to “no” if you don’t want to treat DIN events separately. Thus, using the above two parameters, you can choose whether you want to treat DIN events as combined, separate, both, or completely ignored. |
| − | + | ||
| − | + | ||
| − | Thus, using the above two parameters, you can choose whether you want to treat DIN events as combined, separate, both, or completely ignored. | + | |
| − | + | ||
'''CombineDINeventsPrefix'''<nowiki>=dinComb</nowiki> | '''CombineDINeventsPrefix'''<nowiki>=dinComb</nowiki> | ||
| − | + | This defines the text preceding the number when DIN events are combined. The default is “dinComb”.</div> | |
| − | + | === Harmonie === | |
| − | + | ||
'''Default settings provided for section [Harmonie] (Stellate Harmonie systems):''' | '''Default settings provided for section [Harmonie] (Stellate Harmonie systems):''' | ||
| Line 661: | Line 689: | ||
| − | + | === Neuroscan Keys === | |
| − | + | ||
'''Note that there is a setting "NeuroScanDataNumberOfBits" in the [Defaults] section of BESA.ini that is used for distinguishing the data format of Neuroscan files (16 or 32-bit).''' | '''Note that there is a setting "NeuroScanDataNumberOfBits" in the [Defaults] section of BESA.ini that is used for distinguishing the data format of Neuroscan files (16 or 32-bit).''' | ||
| Line 689: | Line 716: | ||
| − | + | === NKT2100 === | |
| − | + | ||
'''Default settings provided for section [NKT2100] (Nihon Kohden EEG 21xx systems):''' | '''Default settings provided for section [NKT2100] (Nihon Kohden EEG 21xx systems):''' | ||
| Line 714: | Line 740: | ||
| − | + | === Vangard === | |
| − | + | ||
'''AlwaysOpenFileSelect=Yes''' | '''AlwaysOpenFileSelect=Yes''' | ||
| Line 724: | Line 749: | ||
| − | + | === XLTEK === | |
| − | + | ||
'''TriggerScan=Off '''Set to "On" to scan the data file for trigger events | '''TriggerScan=Off '''Set to "On" to scan the data file for trigger events | ||
Revision as of 16:48, 8 April 2024
| Module information | |
| Modules | BESA Research Basic or higher |
| Version | BESA Research 6.1 or higher |
Contents
Introduction
BESA.ini File
BESA Research uses settings provided in the initialization file BESA.ini whenever BESA Research is started or a new file is opened for the first time. The format of this file conforms with standard initialization files (*.ini) of Windows. You may change the settings in BESA.ini using Notepad.exe from the ACCESSORIES group, or other plain text editors to adapt BESA Research to your own everyday needs. The default settings provided in BESA.ini will be used by BESA Research whenever BESA Research or the launch program is started. It is advised that you make a backup copy of BESA.ini before you change the default settings.
Location of BESA.ini
You can place BESA.ini at three possible locations:
- Private: each user on a PC should have his/her own private settings. This is normally in Documents/BESA/Research_7_1
- Public: all users should use one setting, but they can edit BESA.ini to change the settings. This is normally in Public Documents/BESA/Research_7_1
- Administrator: the PC administrator determines the settings. This is normally in C:Program Files(x86)/BESA/Research_7_1
The actual folder names depend on the operating system and the system language.
When BESA starts, it first looks for the administrator version of BESA.ini. If this is not found, it looks for the private version. If this is not found, it looks for the public version. If this is not found, internal default values are used.
There are 13 general sections, and several reader-specific sections:
| [Defaults] | General settings (filters, scaling, and various other settings) |
| [Folders] | Folders used by BESA Research (Examples, Montages, Scripts, Settings,...) |
| [Electrodes] | Electrode renaming |
| [Patterns] | Rename patterns in the Tags menu |
| [Artifacts] | Settings for artifact correction |
| [KEYCONTROLS] | Function key definitions |
| [Search] | Default parameters for search |
| [FFT] | Frequency band definitions |
| [Printer] | Printer control |
| [Calibration] | Calibration control |
| [Video] | Digital video control |
| [Mapping] | Mapping control |
| [Updates] | Options for program updates |
| [Matlab] | Settings for the MATLAB interface |
| [fMRI] | Settings for the fMRI arfifact removal |
| [Montages] | A setting for a default source montage |
Reader-specific settings
[BrainLab]
[Bio-Logic]
[EDF+] [BDF] [Trackit]
[EGI]
[Harmonie]
[NeuroScan Keys]
[NKT2100]
[Vangard]
[XLTEK]
Defaults
Default settings provided for section [Defaults]:
DatabaseAllowLocalFiles=Yes (If set to "Yes", BESA Research will write filenames "datafilename.ftg" and "datafilename.fst" to the data folder, saving current file tag and display settings there. If set to "No", these files are only written to the database. If set to "Yes", you can copy these files along with the data to a new folder, and display settings and tags will be preserved.)
DataBuffering=Off (If set to "On", an internal buffer of length 180 s of data is kept to speed up paging). This can speed up paging, particularly when the data are in a network folder.
DisplayedTime=10 displayed time window [s] on the screen
Montage=Org montage used when opening a new file
ScpScale=50 scale of scalp channels in [mV]
PgrScale=500 scale of polygraphic channels in [mV]
IcrScale=500 scale of intracranial channels in [mV]
MegScale=200 scale of MEG/GRA channels in [fT or fT/cm]
MagScale=1000 scale of MAG channels in [fT] (this feature requires BESA Research 7.1 or higher)
SrcScale=100 scale of source of source montages
BaselineCorrection=On baseline correction, do not switch off in AC systems
ClippingPercent= set from 100 to 200 if you want to clip artifacts in displayed EEG (not used if empty or 0)
LowFilter= low filter cutoff frequency [Hz] (variable filter)
TimeConstant=0.3 time constant for low filter cutoff frequency [sec] (fixed forward filter, 0.3 sec is equivalent to 0.53 Hz)
HighFilter=70 high filter cutoff frequency [Hz] (variable filter)
NotchFilter=50 notch filter center frequency [Hz]
NotchFilterStatus=Off notch filter is off, set=On if you want to use as default
BandFilter=12 band pass filter center frequency [Hz]
BandFilterStatus=Off band pass is off, set=On if you want to use as default
AdditionalChannelFile= defines the full path and name of an additional channels montage file, e.g. C:\Program Files\BESA\Research_x\Montages\AdditionalChannels\EKG.sel
ColoredWaveforms=On scalp waveforms are (not) colored according to region
WriteSegmentPath= defines default path for saving segments/averages. If blank, the path of the current data file is used.
ShowSubjectInfo=Off subject info will (not) be displayed.
ParallelComputing=On defines if parallel computing during extensive computation should be used or not (this feature requires BESA Research 7.1 or higher)
MapSmoothing=0 set a non-zero value to specify a default map smoothing parameter (normally specified in Options/Mapping/Spline Interpolation Smoothing Constant). Valid values are within the range between 1e-8 and 1e-4. Values outside this range will be set to within the range.
The following optional parameters are not defined as default and can be set manually in BESA.ini:
TextEditor="Notepad.exe" defines the path to your preferred text editor. This will be used when you press the Edit button in the Load Coordinate Files dialog box.
NeuroScanDataNumberOfBits=32 defines the format of NeuroScan data files ('16' for 16-bit, '32' for 32-bit). If this variable is not specified, BESA uses a heuristic to (try to) decide which of the two data formats is used. This variable overrides the heuristic. If you want to specify the NeuroScan data format for specific files, create a file, named "16bit" or "32bit", and place it in the data folder.
ScaleAmplitudesForNNChannels=25 Scale waveforms as if a fixed number of channels were displayed in the window (here: 25). A minimum of 10 channels can be used for the scaling. This parameter is superseded if the parameter "ScaleAmplitudesFixedPixelHeight" is specified.
ScaleAmplitudesFixedPixelHeight=70 Set the scale bar for amplitudes to a fixed pixel height (here: 70). If this parameter is set in the .ini file, it supersedes the parameter "ScaleAmplitudesForNNChannels".
Notes
Check the Menu descriptions for the various definitions of filters, montages etc. For montage preselection, use the labels as visible on the montage push-buttons.
The additional channels file should contain all polygraphic channels (e.g. EKG, EOG, respiratory) that you want to view regularly along with the scalp channels. The entry AdditionalChannelFile must specify the full path pointing to the location of additional channel files (recommended: Montages\AdditionalChannels). If no drive is specified, the installation drive of BESA is used.
If BaselineCorrection is set to 'On', before displaying a screen of data, BESA subtracts for each channel the mean over its displayed time points. This optimizes viewing, because it ensures that the vertical position of each channel is not shifted upward or downward from the channel label at the left of the screen. There are some cases in which you will not want baseline correction, i.e. when the DC level in the data is already correctly defined. This is usually the case, for instance, when reading in files that have been processed by BESA. In this case, BaselineCorrection should be set to 'Off', because otherwise maps and source montage displays may be distorted.
Folders
The [Folders] section defines where BESA Research places its files. In versions 5.1 and earlier, files were located in various subfolders of the program folder. This led to problems if the user did not have administrator rights, e.g. to create or write to a file. If you wish, you can also specify paths in the [Folders] section to use the previous locations. The previous location is given for each variable.
These settings allow some flexibility that can be useful if you want to tune BESA Research for use by several users, or on a network. For instance, the Examples and Montages folders might be located on a network disk. For the current defaults, the database, Examples, Montages, and Scripts are set up for use by all users on the PC on which BESA Research is installed. The settings files (Besa.set, Besa.cfg, etc.) are located in private folders so that each user retains his or her own settings.
The default settings (i.e. settings that BESA Research uses if the entries are omitted in the .ini file) are shown for each variable definition.
The folder definitions can use placeholders, labels enclosed by a % sign (e.g. %localapp%), to define paths that vary depending on the language version and on the Windows system. These are defined below.
The Variables
Database=%localapp% The path of the BESA Research database folder (used to be %progdir%System\DB in BESA versions up to 5.1.x). Unless the provided path ends with \DB or \Database, BESA Research will automatically create a folder named Database in the provided path.
Settings=%privatprog%Settings The path of the BESA Research settings folder (used to be %progdir%System in BESA versions up to 5.1.x)
Montages=%publicprog%Montages The path of the BESA Research montages folder (used to be %progdir%Montages in BESA versions up to 5.1.x)
Scripts=%publicprog%Scripts The path of the BESA Research Scripts folder (used to be %progdir%Scripts in BESA versions up to 5.1.x)
Examples=%publicprog%Examples The path of the BESA Research Examples folder (used to be %progdir%Examples in BESA versions up to 5.1.x)
User=%privatprog%Settings The path for user defined settings (used to be %progdir%System\Userdirs in BESA versions up to 5.1.x)
DataExport=%privateprog%Export The path for data to be exported for BESA Connectivity (not listed by default, but can be adjusted by the user)
Placeholders
The strings enclosed by percent signs (%) are placeholders for the following folders in English-language versions of Windows. Folder names differ depending on Windows version, and for other language settings. BESA Research will substitute the placeholders by the appropriate folder name for the system and the system language:
Windows 7, 8.1, and 10 (English):
%localapp% = "C:\Users\[user]\Documents\BESA\Research_7_0", where [user] is the logon name of the current user. This folder is directly accessible from the Desktop as "Desktop\[user]\Documents\BESA\Research_7_0".
%publicprog% = "C:\Users\Public\Public Documents\BESA\Research_7_0".
%privateprog% = "C:\Users\[user]\Documents\BESA\Research_7_0", where [user] is the logon name of the current user. This folder is directly accessible from the Windows Explorer as "Desktop\[User]\Documents\BESA\Research_7_0".
%progdir% = the BESA Research root folder. In a default installation, this is "C:\Program Files (x86)\BESA\Research_7_0".
%besaroot% is the same as %progdir%
Electrodes
This section allows for automatic relabeling of electrodes. For instance, the 10-20 label "T3" can be replaced by the 10-10 convention "T7".
Default settings provided for section [Electrodes]:
T7=T3 replace 10-10 label with old 10-20 convention
T8=T4 replace 10-10 label with old 10-20 convention
P7=T5 replace 10-10 label with old 10-20 convention
P8=T6 replace 10-10 label with old 10-20 convention
X1=ECG1 define X1 channel to be ECG1
X2=ECG2 define X2 channel to be ECG2
Other examples, depending on your electrode input box definition, could be:
PG1=LO1 define X3 as lateral orbital eye electrode left
PG2=LO2 bipolar LO1-LO2 defines horizontal EOG (additional channel)
X3=IO1 infraorbital, e.g. use with FP1 as additional channel for VEOG
X9=Rsp define X9 channel to be a respiratory channel
Relabeling of channel names (as stored in the EEG file header) is helpful to predefine your standard sequence of channels and to avoid the need for reading and/or editing a Channel Configuration file for every EEG file.
Note 1: For polygraphic channels, or if your EKG has been recorded differentially, you should edit and define an Additional Channels Montage according to your recording channel configuration (e.g. Fp1-IO1=vertical EOG). The Additional Channels group permits to display these channels regularly below the scalp montages with individual scales.
Note 2: EOG channels record both eye and scalp activity. In digital EEG systems, EOG electrodes should be labeled according to their position in the 10-10 system (see "Electrode Conventions"). This permits use of these electrodes for mapping and suppression of eye artifacts. The standard definitions above give an example of how to relabel extra channels (X1...X10, PG1, PG2) for the use of EOG, EKG and respiratory (Rsp) channels. Use an Additional Channels file to define horizontal and vertical EOG channels by using the appropriate electrodes in a bipolar montage (an example is provided in eog-ecg.mtg in Montages\AdditionalChannels). Differentially recorded EKG and respiratory channel can be defined in the same file.
Patterns
Default settings provided for section [Patterns]:
These settings define labels for each of the five patterns. The labels are shown* in the Tags menu,
- in the TAG push-button popup menu, and
- when displaying tag info clicking with the right mouse on a tag at the bottom of the EEG or on the event bar.
By default, no labels are defined. Define a label, e.g. for Pattern1 and Pattern2, as in the following example:
Pattern1=Spike
Pattern2=Sharp Wave
Artifacts
Artifact default settings:
See the chapter "Artifact Correction / Reference / Artifact settings in the BESA.ini file" in the online help.
Search
Default settings for pattern search.
Default Settings for the Search/Options Dialog box:
CorrelationThreshold = 75%
AmplitudeThreshold = 100 µV
GradientThreshold = 25
Default Settings for the Search/Average/View (SAV) Dialog box:
PreCursor = -250 ms
PostCursor = 150 ms
HighPassFreq = 2 Hz
HighPassSlope = 12 dB/Octave
HighPassType = 0 (0 = zero phase, 1 = forward, 2 = backward
LowPassFreq = 35 Hz
LowPassSlope = 24 dB/Octave
LowPassType = 0 (0 = zero phase, 1 = forward, 2 = backward)
CorrelationThresholdNoMarked = 60%
Default correlation threshold if no channel labels are marked when the SAV Dialog is opened.
CorrelationThresholdOneMarked = 85%
Default correlation threshold if one channel label is marked when the SAV Dialog is opened.
CorrelationThresholdFourMarked = 65%
Default correlation threshold if between two channel labels are marked when the SAV Dialog is opened.
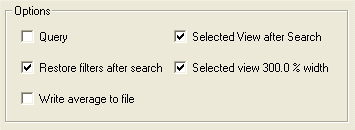
SelectedViewWindowWidthMultiplier = 300%
WriteAfterSearch = No
If set to "Yes", a File Save dialog will open, to allow to save the search average to a file (as with the SAW function).
WriteAfterSearchCheckBox = No
If set to "Yes", an additional checkbox "Write after search" is displayed at the bottom of the SAV Dialog, allowing to choose whether or not to write the search average after a search:
PreserveDefaults = Yes
If set to "No", the SAV Dialog will open with the same boxes checked as the last time the dialog was opened during the current session.
If set to "Yes", the default frequency, buffer width, selected view after search, and default threshold are always checked when the dialog is opened.
KeyControls
In the [KeyControls] section you can specify functions that can be allocated to function keys or to the Del key. Specify using the form:
Fn=function or
Del=function
where "n" is a number between 2 and 12 (F1 is reserved for Help). For example:
F2 = Batch1
Possible functions are:
Setting or removing events:
Patternn, where n=1-5: Sets the tag number n at the cursor latency.
Epochfast: sets one boundary of an epoch at the cursor latency, but does not open the epoch text box to define a label.
Marker: sets a marker at the cursor latency.
Comment: sets a comment at the cursor latency and opens the comment box to enter text.
Epoch: sets one boundary of an epoch at the cursor latency and opens the epoch text box to enter a label.
Artifact: sets one boundary of an artifact segment at the cursor latency.
Delete: deletes a tag at the cursor latency
Batches and Montages:
Batchn, where n=1-12: Runs a predefined batch file corresponding to the number n.
Montagen, where n=1-12: Sets a montage corresponding to the number n.
The default settings after program installation are listed in the online help chapter Review / Reference / Controls / Mouse and Keyboard / Keyboard Controls.
FFT
Default settings provided for section [FFT]:
These settings define the setup in the Spectral Analysis section of the BESA Research program (FFT window, see the chapter "Spectral Analysis / FFT"). Up to 7 frequency bands may be defined. Five are defined by default.
FFTBand1=On FFT Bands 1-5 are defined
FFTBand2=On
FFTBand3=On
FFTBand4=On
FFTBand5=On
FFTBand6=Off FFT Bands 6-7 are not defined
FFTBand7=Off
FFTNameBand1=Delta Names of the defined bands
FFTNameBand2=Theta
FFTNameBand3=Alpha
FFTNameBand4=Beta
FFTNameBand5=Gamma
FFTColorBand1=RGB(0,0,0) Default color of each band
FFTColorBand2=RGB(0,128,64)
FFTColorBand3=RGB(128,0,0)
FFTColorBand4=RGB(255,0,0)
FFTColorBand5=RGB(255,128,0)
FFTColorBand6=RGB(255,192,0)
FFTColorBand7=RGB(255,255,0)
FFTLowBand1=1 Delta from 1-4 Hz
FFTHighBand1=4
FFTLowBand2=4 Theta from 4-8 Hz
FFTHighBand2=8
FFTLowBand3=8 Alpha from 8-14 Hz
FFTHighBand3=14
FFTLowBand4=14 Beta from 14-30 Hz
FFTHighBand4=30
FFTLowBand5=30 Gamma from 30-50 Hz
FFTHighBand5=50
These values are best set from within BESA Research, using the Options menu in the FFT window (see the chapter "Spectral Analysis / FFT / FFT Options Menu"). Current settings are stored after each session and retrieved in the next session.
Printer
Default settings provided for section [Printer]:
PrinterMarginPercent=100 controls size of printout
PrinterColors=256 set to 1/2 for black&white, 0/256 for color printers
PrinterLineMode=1 set to 2 for thicker lines and to save printer memory
PrinterMapResolution=1 set to 2, 3, 4 to save printer memory and increase speed
Calibration
Default settings provided for section [Calibration]:
AutoCalibration=Off On: automatic calibration of signals >= 4 cycles
MicrovoltCalibration=50 peak voltage of calibration signal
If calibration is set to On, the menu item Calibration will appear in the Process menu. Position your current screen at an epoch containing at least 4 regular cycles of the calibration signal (in all channels!) and select Calibration.
Video
Default settings provided for section [Video]:
DVCFilePath=C:\DVC\DVPlay.exe holds the path to the digital video player
DVCCommandLineArguments=/S:3 /M:P /T:M arguments to be passed to the digital video player
CursorPagingOffsetLeft=0.2
CursorPagingOffsetRight=0.8
CursorMinDistToBorderBeforePaging=0.02
PageDisplayIfCursorIsBelowVideo=1
MappingRepetitionRateWithVideoInMS=100 gives the number of milliseconds between two maps if the mapping window is open while the video is running. If the graphics board encounters problems during the display, this value should be increased.
Mapping
Default settings provided for section [Mapping]:
UseBitmapDrawing=Off
Set this to "On" if 3D maps show a strange pattern of black triangular shapes (this is frequently observed with modern Intel On-Board graphics controllers, and is a result of inadequate drivers for OpenGL).
Use3DVBlending=Auto
Set this to "Off" if the 3D view in the Montage Editor or the Source Analysis window does not show up properly (this may happen with some older graphics cards).
Set this to "On" if the 3D view in the Montage Editor or the Source Analysis window shows a ragged surface boundary.
UseDoubleBuffering=On
Set this to "Off" to disable double buffering mechanism that prevents the screen from flickering while paging through data and dragging window (this feature requires BESA Research 7.1 or higher).
Note: MapSmoothing, the default map smoothing parameter, can be specified in the [Defaults] section.
Matlab
Default settings for the [Matlab] section:
Platform=64
Set Platform=32 if you want to use the x86 version of MATLAB.
Updates
This section is not normally required, but the variables here can be altered or defined to determine how BESA Research checks for dongle and program updates.
DaysBetweenUpdateChecks=7
Sets the number of days between automatic checks for updates. Set the value to 0 to check every time BESA Research is started. Set to -1 to turn off automatic update checks.
CheckNetworkDongle=Off
For the network administrator: If set to "On", BESA Research will check the dongle on the network for updates. Otherwise the state of the network dongle will be ignored.
LocalPath
For the network administrator. This can be set to a path on the local network to the BESA update files, so that users can obtain their updates locally. The path is given to the text file "UpdateVersions.txt" (e.g. LocalPath=\\transtec-sak\zarascratch\BESA\Updates\UpdateVersions.txt), which contains further details for the program to obtain its updates. If you want to use this feature, please contact us using our support portal.
The following variables are not required, because BESA Research has the paths hardwired:
FTP1 (also FTP2, FTP3)
Download server
Path1 (also Path2, Path3)
Path on the server to UpdateVersions.txt.
HaspPath1 (also HaspPath2, HaspPath3)
Path on the server to HASP (dongle) update files.
History
Path on the server to general history file
FMRI
(requires Besa Research 7.0 or higher)
These settings define the default parameters for the fMRI artifact removal in the BESA Research (see fMRI artifact removal chapter for further details). For example:
[FMRI] FMRIRemovalMode=1 TRDelay=200 TRLength=800 NumberOfAverages=21 fMRImoveThreshold=0.15 FMRITRID=8015 ScansToSkip=0
These values indicate:
- FMRIRemovalMode: Removal method (0: Turned off; 1: Allen et al, 2000; 2: Allen et al., 2000 Modified; 3: Moosmann et al.,2003)
- TRDelay: Delay between marker and start of volume acquisition [ms]
- NumberOfAverages: Number of artifact occurrence averages
- fMRImoveThreshold: Movement threshold [mm]
- FMRITRID: fMRI Trigger code
- ScansToSkip: Number of scans to skip
Montage
The section [Montage] allows to specify an initial montage that is set the first time when the source (Src), recorded (Rec), virtual (Vir) or user (Usr) montage button is pressed. If BESA.ini does not specify a montage, pressing the corresponding button opens the drop-down menu offering all the available montages for the current montage type.
Source=25s specifies that when the Src button in the control ribbon is pressed for the first time, the source montage "25s" will be selected.
Recorded=Original Recording specifies that when the Rec button in the control ribbon is pressed for the first time, the source montage "Original Recording" will be selected.
Virtual=Triple Banana specifies that when the Vir button in the control ribbon is pressed for the first time, the source montage "Triple Banana" will be selected.
User=CA25 specifies that when the Usr button in the control ribbon is pressed for the first time, the source montage "CA25" will be selected.
Reader-Specific Settings
BrainLab
Default settings provided for section [BrainLab]:
BrainLabFormat=New this entry ensures that the newer BrainLab file format can be read by BESA Research.
Bio-Logic
FileSelect=Yes
If there are several Bio-Logic files in a data folder, the reader can check if the files have the same settings. There are three possible options:
- Open a dialog to ask if the files should be treated as a single data set, or as individual, separate files.
- Always concatenate such files into a single data set. In this case use FileSelect=All
- Always open the files as single, separate files. In this case use FileSelect=Single
EDF+/BDF/Trackit
TriggerScan=On
Set TriggerScan=Off to prevent BESA Research from scanning the file for triggers. This is done separately for EDF+, BDF, and Trackit files in sections [EDF+], [BDF], and [Trackit] in the BESA.ini file.
EGI
The treatment of DIN events can be modified in the [EGI] section:
CombineDINevents=yes/no (default is “yes”)
Set to “no” if you want to treat DIN events separately, and not generate combined values.
SeparateDINevents=yes/no (default is “yes”)
Set to “no” if you don’t want to treat DIN events separately. Thus, using the above two parameters, you can choose whether you want to treat DIN events as combined, separate, both, or completely ignored.
CombineDINeventsPrefix=dinComb
This defines the text preceding the number when DIN events are combined. The default is “dinComb”.</div>
Harmonie
Default settings provided for section [Harmonie] (Stellate Harmonie systems):
SeizurePreEpoch=60 length of the epoch preceding a seizure detection in s
SeizurePostEpoch=60 length of the epoch following a seizure detection in s
PushButtonPreEpoch=60 length of the epoch preceding a push button detection
PushButtonPostEpoch=60 length of the epoch following a push button detection
When BESA Research encounters a seizure detection event or a push button detection event in a Stellate Harmonie file, it automatically sets an epoch around the event, which makes it convenient to view just those epochs for analysis. The length of the epochs preceding and following the events can be adjusted in the .ini file.
Neuroscan Keys
Note that there is a setting "NeuroScanDataNumberOfBits" in the [Defaults] section of BESA.ini that is used for distinguishing the data format of Neuroscan files (16 or 32-bit).
Default settings provided for section [NeuroScan Keys] (NeuroScan systems):
Event1=Movement Text corresponding to keyboard events 1 through 10
Event2=Blink
Event3=Talking
Event4=Cough
Event5=Muscle
Event6=Jaw
Event7=Sneeze
Event8=Swallow
Event9=Eye movement
Event10=Hiccup
NKT2100
Default settings provided for section [NKT2100] (Nihon Kohden EEG 21xx systems):
TriggerScan=On Set to "Off" to prevent a scan for trigger events.
Country=NotKanji set to NotKanji for non-Kanji characters else to Kanji
KanjiCharSize=16 Kanji character size
KanjiPrinterCharSize=32 Kanji printer character size
EEG_Sensitivity=50 default sensitivity of Nihon Kohden EEG-2100 system
DC_Sensitivity=50 default sensitivity of Nihon Kohden DAE-2100 system
QJ_Sensitivity=100 default sensitivity of Nihon Kohden QJ-403 system
Mark_Sensitivity=100 default sensitivity of EEG-2100 marker channels
These settings need to be changed only if the manufacturer has specified different gains for your system. Otherwise do not alter these settings.
Vangard
AlwaysOpenFileSelect=Yes
If "Yes" is selected, each time a Vangard file is opened, a dialog box will open, asking for a selection of the segment type to display.
If "No" is selected, the selection dialog is opened whenever a Vangard file is opened for the first time, or if the Channel and digitized head surface point information dialog box is opened (e.g. with ctrl-L or File/Head Surface Points and Sensors/Load Coordinate Files... ).
XLTEK
TriggerScan=Off Set to "On" to scan the data file for trigger events
MontageNo=2 Set to 1 or 2. If two montages for the data file are defined, this variable determines whether the first or the second alternative should be used.
| Review | |
|---|---|
| Source Analysis | |
| Integration with MRI and fMRI | |
| Source Coherence | |
| Export | |
| MATLAB Interface | |
| Special Topics |
|