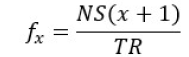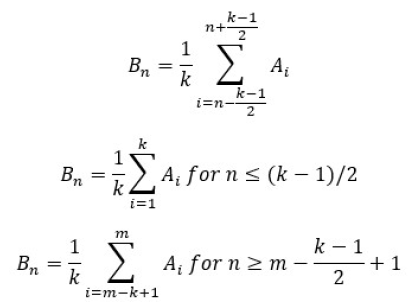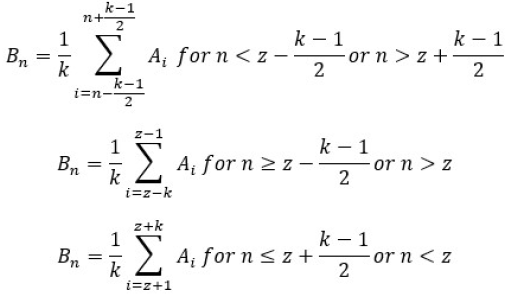Difference between revisions of "BESA Research Artifact Correction"
(→References) |
|||
| (37 intermediate revisions by 5 users not shown) | |||
| Line 1: | Line 1: | ||
| − | == | + | {{BESAInfobox |
| + | |title = Module information | ||
| + | |module = BESA Research Basic or higher | ||
| + | |version = 6.1 or higher | ||
| + | }} | ||
| − | === | + | <!-- == Artifact Correction == --> |
| − | + | == Introduction == | |
| − | + | BESA Research includes a sophisticated spatial filtering method to detect and correct artifacts. The method can be applied to any kind of artifact including EOG and EKG artifacts. Spatial topographies describing the artifacts may be defined manually or by allowing the program to scan the data automatically. Manual definition can be accomplished by marking a time range containing the artifact or a buffer with the averaged artifact. In particular, EKG artifacts should be averaged before defining the spatial topography. Once defined, the corrected data waveforms can be displayed, together with additional channels below the current montage showing the time course of the artifacts. Spatial artifact topographies can be saved to a file (<span style="color:#ff9c00;">'''<nowiki>*.atf</nowiki>'''</span>), and may be recalled for later use with the same data or other data from the same subject. | |
| − | + | Automatic artifact correction is described in the chapter Automatic Correction below. For a tutorial on using calibration data for an accurate estimation of artifact coefficients, see help chapter ''"Artifact Correction / Tutorial".'' | |
| − | |||
| + | '''How correction works''' | ||
| − | + | Artifact correction is based on the spatial components method for correcting eye artifacts (Berg and Scherg 1994, see also Ille et al. 1997, 2002). Artifact and brain signal subspaces are described in terms of spatial topographies. The correction process consists of four steps: | |
| − | + | ||
| − | + | ||
| − | + | ||
| + | # Define the topography for each type of artifact using automatic detection or manual definition | ||
| + | # Determine the brain signal topographies underlying the displayed EEG segment | ||
| + | # Reconstruct the artifact signal at each scalp electrode with a spatial filter taking into account artifact as well as brain signal subspace | ||
| + | # Subtract the reconstructed artifact signal from the original EEG segment | ||
| − | |||
| − | + | '''Available methods for correction''' | |
| − | + | BESA Research allows artifact correction to be done in different ways. The method you choose depends on how you want to analyze your data. The methods vary in how the data is distorted by correction, and how the brain activity is modeled for the estimation of eye activity: | |
| − | * | + | * '''Artifact-free review of the ongoing EEG'''. Use the '''PCA method''' (Ille et al. 2002). This is optimized to contrast the artifacts against the predominant rhythmic EEG activities. |
| − | * | + | * '''Analysis of averaged data waveforms (ERPs).''' Use the '''Surrogate method''' (Berg and Scherg 1994). Brain activity is modeled by a fixed dipole model (the "surrogate model"). Data waveforms will be distorted least if the Surrogate model is close to the dipole model of the averaged data. |
| − | * | + | * '''Source analysis'''. Use the '''Optimizing method''' (Berg and Scherg 1994). This is available in two variants: |
| + | ** '''Develop dipole models in the presence of spatial components'''. This is the preferred method. Create separate averages of the ''uncorrected'' conditions and of the artifact events, e.g. blinks. All averages can be concatenated into one binary segment file (<span style="color:#ff9c00;">'''<nowiki>*.fsg</nowiki>'''</span>). The averaged artifacts should then be used to define spatial components during source analysis. Thus, the time course of the artifacts and of the event-related source activity can be separated and observed in the source analysis window. | ||
| + | ** '''Develop dipole models on the pre-corrected averages'''. Using subspace-projection (SSP), the artifact dimensions are compensated for in source models. This will lead to the same dipole models as above, but a) the time course of artifact activity is not available, and b) residual variance is lower because variance from dimensions in data space occupied by the artifact spatial components are removed from the data by SSP. It does not matter which correction method was used with BESA Research (PCA, Surrogate, or SSP) – the results of source analysis are the same. | ||
| + | * '''No artifact correction'''. If the data contain only a few artifacts, use artifact rejection rather than correction. This avoids the distortion resulting from correction. | ||
| − | |||
| − | |||
| − | + | '''Artifact correction in BESA Research distorts the data less than traditional eye artifact correction''' | |
| + | Artifact correction can distort the data topographies and reduce or augment activities at some scalp regions, e.g. frontal scalp after eye movement correction. Therefore, maps should be interpreted with caution. | ||
| − | + | The advantage of the methods used in BESA Research over traditional approaches is that overlapping contributions of the artifacts and the EEG are separated as much as possible. In traditional approaches, a proportion of the signal on an EOG channel is always subtracted from each EEG signal. Thus, in contrast to traditional methods, the topography of the EEG is less distorted by correction based on spatial components, even if the correlation between artifact and brain activity is high. | |
| − | |||
| − | + | '''Source Analysis takes distortion due to artifact correction into account''' | |
| + | However much the signals may be distorted by artifact correction, the distortion can be taken into account in Source Analysis. This is accomplished using signal subspace projection (SSP, cf. Uusitalo et al. 1997), which is equivalent to the "optimizing" correction method described in Berg and Scherg (1994). | ||
| − | + | SSP is switched on in Source Analysis: | |
| − | <span style="color:# | + | * if data were averaged with artifact correction switched on, and the averaged data were saved in BESA Research binary format (<span style="color:#ff9c00;">'''<nowiki>*.fsg</nowiki>'''</span>). |
| + | * if artifact correction is switched on for any other type of file. For instance, you may have averaged a file to ASCII format. Read in the file, and load the artifact coefficients. Make sure correction is switched on. | ||
| − | + | The PCA and Surrogate methods distort less than traditional EOG subtraction methods, but they do not eliminate distortion! If you do not use the first variant of the Optimizing method in source analysis, you should make sure that subspace projection (SSP) is switched on in the source analysis module. This is indicated by the text "ART" at the right of the status bar in the source analysis module. Visual feedback that SSP is switched on is supplied in the Source Module by the text ART at the right of the Status Bar: | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| Line 58: | Line 58: | ||
| − | + | '''References''' | |
| − | + | * Berg, P., and Scherg, M. A multiple source approach to the correction of eye artifacts, Electroenceph. clin. Neurophysiol., 1994, 90: 229-241. | |
| − | + | * Ille N., Berg P., and Scherg M. A Spatial Components Method for Continuous Artifact Correction in EEG and MEG. Biomed. Tech., 1997, 42 (suppl. 1): 80-83. | |
| − | + | * Ille, N., Berg, P., Scherg, M. Artifact correction of the ongoing EEG using spatial filters based on artifact and brain signal topographies. J. Clin. Neurophysiol. 2002, 19: 113-124. | |
| − | + | * Uusitalo, M.A., Ilmoniemi, R.J. Signal-space projection method for separating MEG or EEG into components. Med. Biol. Eng. Comput., 1997, 35: 135-140. | |
| − | + | '''Artifact Correction Menu''' | |
| − | + | In the Review Window use the <span style="color:#3366ff;">'''Artifact'''</span> menu (<span style="color:#3366ff;">'''Alt+A'''</span>) to: | |
| − | + | [[Image:artifact_menu.png]] | |
| + | * assign EOG or ECG artifact topographies automatically | ||
| + | * assign artifacts manually | ||
| + | * display the reconstructed time course of the artifacts below the EEG | ||
| + | * display the EEG with the artifacts removed | ||
| + | * correct artifacts caused by simultaneous fMRI recording | ||
| + | * delete assigned artifacts | ||
| + | * select which artifact topographies to use | ||
| + | * display maps of the topography defining each artifact | ||
| + | * set options for modelling the brain signal subspace | ||
| + | * load a previously defined set of artifact topographies | ||
| + | * save the current set of artifact topographies to a file | ||
| − | + | == Automatic Artifact Correction == | |
| − | + | BESA Research provides an automatic artifact correction method that enables users to quickly obtain artifact-cleared data. For more details see the main article on [http://wiki.besa.de/index.php?title=BESA_Research_Automatic_Artifact_Correction Automatic Artifact Correction] | |
| − | + | == Using Defined Topographies for Correction == | |
| − | + | It is often desirable to use real data topographies for artifact correction, since they reflect the individual variability, e.g. in eye artifacts. The easiest way to assign an individual topography is to mark a data block which contains an artifact, and define the topography via the right click popup menu. The proceeding is as follows: | |
| − | + | * Mark the artifact in the display window using the left mouse drag: | |
| − | + | [[Image:Artifact Correction (7).gif]] | |
| − | |||
| − | < | + | * Select the menu item <span style="color:#3366ff;">'''Artifact / Select'''</span>. Alternatively, right click and select ''Define Topography'' from the popup menu which appears: |
| − | + | [[Image:Artifact Correction (8).gif]] | |
| − | |||
| − | + | * The ''Select Topographies'' dialog box appears. There is a list of artifact types, for which topographies can be assigned. To assign a blink artifact, click into the check box in the third row. The topography is activated, and a map of the artifact topography is shown automatically: | |
| − | * | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| + | [[Image:Artifact Correction (9).gif]] | ||
| − | + | ''EEG data after a topography has been assigned to the category "Blink". A map of the topography is shown at the right. An additional channel for the blink artifact is introduced at the bottom, and data are corrected automatically.'' | |
| − | |||
| − | <span style="color:#3366ff;"> | + | Note that the data are automatically corrected, and an additional virtual artifact channel appears at the bottom. To switch correction off again, use the menu item <span style="color:#3366ff;">'''Artifact / Correct'''</span> which toggles correction. |
| − | + | The artifact topography can be estimated with higher accuracy if a number of artifact epochs are averaged into a data buffer. This is especially recommended for correcting EKG artifacts, since the share of the overall topography which is held by the brain activity is larger for EKG artifacts. To average a number of artifacts: | |
| − | + | # Select the channel where the artifact shows most prominently by a left click on the channel label. | |
| + | # Mark the artifact in the data display. | ||
| + | # Press the <span style="color:#3366ff;">'''SAW'''</span> button at the top of the display window: | ||
| − | |||
| − | + | [[Image:Artifact Correction (10).gif]] | |
| + | ''Averaging over an artifact'' | ||
| − | |||
| + | This starts an automatic average of all matching patterns in the data file. You may stop the search after a number of events were found (progress is shown in the status bar; stop averaging with the <span style="color:#3366ff;">'''ESC'''</span> key). When asked for a filename to save the pattern, press <span style="color:#3366ff;">'''Cancel'''</span>. The averaged data buffer containing the artifact is shown. Now use the averaged artifact to define the artifact topography as described above for the blink artifact. When selecting the artifact category, choose EKG: | ||
| − | + | [[Image:Artifact Correction (11).gif]] | |
| + | ''Defining a topography for an EKG artifact from an averaged EKG pattern'' | ||
| − | |||
| − | + | For the correction of EKG artifacts, it is recommended to specify a (bipolar) polygraphic EKG channel. If only scalp channels are available, the correction can work reasonably only if more than 30 channels are available and if inferior electrodes are included. | |
| − | + | Some artifacts can contain more than one spatial component (e.g. EKG artifacts). It is possible to select more than one component for correction. | |
| − | + | More information on artifact correction is given in the help chapter "''Artifact Correction / Tutorial''". | |
| − | + | ==fMRI artifact removal== | |
| − | + | ''(requires Besa Research 7.0 or higher)'' | |
| − | + | ===Introduction=== | |
| − | < | + | <u>Currently data collected during simultaneous EEG-fMRI is not intended to be used in any diagnostic procedures. Following these guidelines, fMRI artifact removal implemented in BESA Research may also not be used for any diagnostic procedures This tool was developed only for research purposes. |
| − | + | Please use only EEG hardware that is certified for MRI!</u> | |
| − | |||
| − | + | When EEG data is recorded during an fMRI session there are plenty problems to solve to be able to analyze the real brain signal The first one is related to fast switching of MRI gradient coils which is a part of volume acquisition in the Echo-Planar sequence used for fMRI imaging This switching introduces variable magnetic fields that induce variable electrical fields in the EEG recording system (electrodes and wires namely), as described by Maxwell’s second law The induced artifact is typically very large in amplitude (typically 16 mV in a 3.0 T MR scanner). Also, the frequency range of the artifact starts in the frequency band of the EEG signal The artifact frequency and its harmonics can be easily assessed using the following equation (Rusiniak et al, 2013b): | |
| − | |||
| − | + | [[Image:FMRI_formula2.png]] | |
| − | |||
| − | < | + | where f<sub>x</sub> is the harmonic frequency, x is the harmonic number, NS represents the number of slices, and TR stands for Repetition Time of fMRI volume acquisition. From a variety of methods, the most relevant successful and best known are those based on moving average subtraction as firstly introduces by Allen and colleagues (2000). These methods work only when the following assumptions are true: |
| − | + | #Each artifact occurrence is identical: Each fMRI volume acquisition is identical (gradient switchingdoes not alter between volumes) so in general all artifacts are identical However, if the subject moves during the acquisition the artifact changes because electrode positions in relation to the local magnetic field have changed. | |
| + | #EEG signal is stationary: In principle this assumption is true for the EEG signal. The signal stationaritycan be affected by the paradigm, therefore jitter between triggers is strongly advised. | ||
| + | #Each start points of artifacts have to be precisely indicated: This can be achieved either by hardwareclock synchronization between EEG system and MRI machine or by a very high sampling rate (>10 kHz). All modern EEG hardware units designed for simultaneous EEG-fMRI registration are capable of clock synchronization. | ||
| − | |||
| − | + | ===Parameters=== | |
| − | |||
| − | [[Image: | + | [[Image:FMRI_articact_removal1.png|900px]] |
| − | + | * '''fMRI Trigger code''' The trigger code will be automatically detected in most cases If not please adjust this value to the correct code of trigger related to fMRI acquisition. | |
| + | * '''Number of scans to average (k)''' Definition of how many surrounding artifact occurrences will be used for template creation (the larger the number, the more stable the artifact template will be, but in a dataset where there is a lot of subject movement the template will be far from good). | ||
| + | * '''Number of scans to skip''' This parameter indicates how many volumes from the beginning of the fMRI recording have to be skipped This setting is rarely used Use it only when scans that are not part of the actual fMRI sequence (i.e. dummy scans) have trigger events assigned that are visible in the EEG recording. | ||
| + | * '''Delay between marker and start of volume acquisition''' In the situation that volume acquisition starts before or after trigger occurrence this value can be used to adjust filtration. | ||
| + | * '''Length of fMRI volume (t)''' This value has to be equal to the fMRI sequence parameter – repetition time (TR) When a new file is opened this value is automatically computed assuming continuous EEG recording (fMRI volumes are recording without any gap). | ||
| + | * '''Movement threshold''' – This setting is only applicable for the last two filtration methods It indicates the minimum acceptable value of movement during the fMRI session. | ||
| + | * '''Realignment file''' A file that is the outcome of the step named “realignment” during fMRI data postprocessing BESA Research accepts files from SPM, BrainVoyager, AFNI, FSL The number of volumes in the realignment file has to match the number of triggers in the EEG data, and only then the last two removal methods will be available In rare cases use '''Number of scans to skip''' to adjust this value to match number of triggers between EEG and fMRI realignment file. | ||
| + | * '''Removal method''' In BESA, three methods are available (see below for more details) The last two methods are available only if a correctly configured '''realignment file''' is provided. | ||
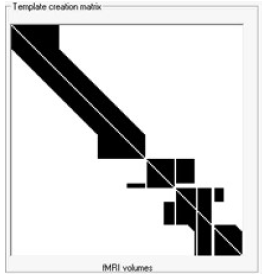
| + | * '''Template creation matrix''' Visualization matrix that illustrates how the filtration is performed Check Filtration Methods description below for further details. | ||
| − | |||
| + | ===Filtration Methods=== | ||
| − | + | ==== Allen et al, 2000 ==== | |
| − | + | This method is a starting point for the two other methods as well In all methods a moving average artifact subtraction is used and the difference between methods can be noted in the template creation matrix | |
| + | To obtain artifact clean data (C) at epoch n the artifact template (B) has to be subtracted from artifact contaminated data (A) | ||
| + | The mathematical principle for creating an artifact template at n-th epoch (Bn) is: | ||
| − | |||
| − | + | [[Image:FMRI_Filtration_Methods1.png]] | |
| − | |||
| − | + | , where ''m'' is the number of fMRI volumes recorded, and ''k'' is the number of artifact volumes used for template creation (averaging). | |
| + | After the artifact template for every epoch is created the final subtraction can be performed: | ||
| − | < | + | :::'''C<sub>n</sub> = A <sub>n</sub> - B<sub>n</sub>''' |
| − | + | This procedure can be explained visually: | |
| − | |||
| − | + | [[Image:FMRI_Filtration_Methods33.png]] | |
| − | |||
| − | + | To clean the epoch indicated by the green shaded box the surrounding epochs have to be averaged first (here the averaging constant is k=10) After the template is created, it is subtracted from the recorded data at each channel independently to obtain artifact free data Next, this procedure is repeated for every epoch related to fMRI acquisition as indicated in the template creation matrix: | |
| − | |||
| − | + | [[Image:FMRI_Filtration_Methods44.png]] | |
| − | |||
| − | + | Note how the template changes when the number of artifact occurrence used for averaging is increased to k=30: | |
| − | |||
| − | + | [[Image:FMRI_Filtration_Methods55.png]] | |
| − | |||
| − | + | ==== Allen et al, 2000 Modified ==== | |
| − | |||
| + | This method is available only if a correctly configured realignment file is provided Also, the changes in filtration will be visible if there is significant movement in the data (a recommended value for the movement threshold is 0.3 mm) In the modified Allen method, the epochs where subject’s movement is exceeding the threshold are ignored during artifact template creation, as shown in the template creation matrix below Note the vertical white lines – they represent the moments were movement was higher than threshold The data will be filtered but less artifact occurrences will be used for template creation close to a movement area. | ||
| − | |||
| − | + | [[Image:FMRI_Filtration_Methods66.png]] | |
| − | |||
| − | + | ==== Moosmann et al, 2009 ==== | |
| + | As with the modified Allen method, this method (Moosmann et al, 2009) is also available only if a correctly configured realignment file is provided. The adjustment of the template creation matrix is much more advanced and can be explained by the following equation | ||
| − | |||
| − | + | [[Image:FMRI_Filtration_Methods77.png]] | |
| − | |||
| − | + | , where all parameters are defined as above and z means an epoch number where movement exceeds the threshold This operation results in the following template creation matrix: | |
| − | |||
| − | + | [[Image:FMRI_Filtration_Methods88.png]] | |
| − | |||
| − | + | Note that this operation results in a different shape of matrix Importantly there are also white vertical lines This indicates that there was more than one epoch contaminated with movement In such situations, the matrix is transformed to adjust to fit to the largest movements, and smaller movements are treated as in Allen et al, 2000 Modified method. | |
| − | + | ===Good Practice=== | |
| − | + | Please keep in mind that simultaneous EEG-fMRI recording is a difficult yet powerful technique Following these rules could help one to perform a successful experiment: | |
| − | + | *For an ERP experiment remember to design it with jitter between trials (i.e. random value of ±200 ms) Also, applying a pure EEG or pure fMRI paradigm will probably have the effect that one of the modalities will not show satisfying results Proper paradigm preparation is essential for success Some further guidelines can be found here:(Rusiniak et al, 2013a) | |
| + | *Inform your subject how important it is not to move during the acquisition. | ||
| + | *Keep electrode to skin impedance as low as possible. | ||
| + | *The EEG-fMRI recording session should be long enough to allow for proper artifact creation Usually an experiment should last at least 6 minutes. | ||
| + | *At the same time try to limit time of experiment to a minimum and preferably perform EEG-fMRI registration before other sequences to limit movement due to inconvenient supine position. | ||
| + | *Place subject about 4 cm further in caudal direction to reduce artifacts The MRI laser crosshair should point not to the Nasion position but to the middle of the forehead (Mullinger et al, 2011) | ||
| + | *Especially for the first few registrations repeat the experiment outside of the bore to compare results | ||
| + | *Remember that conditions in MRI are far from standard EEG recording The long supine position, electrode pressure on back of the head, huge noise and stress can affect the EEG! | ||
| − | + | ===Balistocardiogram and other artifacts=== | |
| − | + | After removing the fMRI gradient artifact many different steps can be performed (Abreu et al, 2018) For ECG induced artifacts (balistocardiogram) we recommend artifact correction using defined topographies for correction (usually with more than 3 components to filter). | |
| − | + | ===References=== | |
| − | + | *Abreu, R, Leal, A, Figueiredo, P, 2018 EEG-Informed fMRI A Review of Data Analysis Methods Front Hum Neurosci 12, 29 https://doi.org/10.3389/fnhum.2018.00029 | |
| + | *Allen, P.J., Josephs, O, Turner, R, 2000 A Method for Removing Imaging Artifact from Continuous EEG Recorded during Functional MRI. NeuroImage 12, 230–239 https://doi.org/10.1006/nimg.2000.0599 | ||
| + | *Moosmann, M, Schönfelder, V.H., Specht, K, Scheeringa, R, Nordby, H, Hugdahl, K, 2009. Realignment parameter-informed artefact correction for simultaneous EEG–fMRI recordings. NeuroImage 45, 1144–1150 https://doi.org/10.1016/j.neuroimage.2009.01.024 | ||
| + | *Mullinger, K.J., Yan, W.X., Bowtell, R, 2011. Reducing the gradient artefact in simultaneous EEG-fMRI by adjusting the subject’s axial position. NeuroImage 54, 1942–1950, https://doi.org/10.1016/j.neuroimage.2010.09.079 | ||
| + | *Rusiniak, M, Lewandowska, M, Wolak, T, Pluta, A, Milner, R, Ganc, M, Włodarczyk, A, Senderski, A, Śliwa, L, Skarżyński, H, 2013a. A modified oddball paradigm for investigation of neural correlates of attention a simultaneous ERP–fMRI study Magn Reson Mater Phys Biol Med 26, 511–526 https://doi.org/10.1007/s10334-013-0374-7 | ||
| + | *Rusiniak, M, Wolak, T, Lewandowska, M, Cieśla, K, Skarzynski, H, 2013b. The relation between EPI sequence parameters and electroencephalographic data during simultaneus EEG-fMRI registration an initial report, in ESMRMB 2013 Congress, Book of Abstracts, Saturday Presented at the ESMRMB, Springer, Toulouse, p 661 https://doi.org/10.1007/s10334-013-0384-5 | ||
| − | + | == Artifacts Tutorial: Using Calibration Data to Generate Artifact Coefficients == | |
| − | + | In this example, we will work on calibration data containing systematic eye movements: | |
| − | + | * Each type of eye movement (left, right, up, down, blink) will be averaged separately, using pattern search. | |
| + | * Correction coefficients for horizontal and vertical eye movements and blinks will be generated. | ||
| + | * We will obtain feedback about the coefficients: how well do these coefficients correct the calibration data? | ||
| + | * Using the PCA in the source analysis module, we can demonstrate the presence of rider artifacts (small eyelid movements) overlapping the saccades. | ||
| − | |||
| − | < | + | The data example, <span style="color:#ff9c00;">'''Examples\Xtras\Eye Artifacts\calibration.foc'''</span>, was collected using 64 channels referred to Cz. In the eye movement calibration procedure, the subject watched a fixation point on a video monitor. At intervals of 2 s, an arrow appeared on the screen. The subject was instructed to move his eyes in the direction of the arrow to a point (marked on the wall of the shielded cabin) approximately 15° away from the fixation point, and then back to the fixation point. The subject was also instructed to try not to blink during the eye movement, but was allowed to blink while watching the fixation point. There were 20 movements in each of the directions left, right, up, down. Following these movements, the arrows were replaced by an image of an eye. This was a cue to blink. 20 eyeblinks were requested. |
| − | + | In the data file, trigger 1 denotes the horizontal arrows, trigger 2 the vertical arrows, and trigger 3 the eyeblinks. | |
| − | + | '''Step 1: Setting up the parameters''' | |
| + | * Load the file, <span style="color:#ff9c00;">'''Examples\Xtras\Eye Artifacts\calibration.foc'''</span>. | ||
| + | * '''Set the ''Default Block Epoch'' to the range -100 ms to +400 ms (Edit/Default Block Epoch).''' | ||
| + | * Select pattern search (<span style="color:#3366ff;">'''Search/Pattern'''</span>), and make sure that the Query option is switched on (<span style="color:#3366ff;">'''Search/Query'''</span>). | ||
| + | * Select buffer 1 for averaging (<span style="color:#3366ff;">'''Tags/Pattern 1'''</span>). | ||
| + | * Set the filters to 0.5 Hz high pass, 6 db/octave, forward, and 8 Hz low pass, 12 db/octave, zero phase (<span style="color:#3366ff;">'''Filters/Edit Filter Settings'''</span>). | ||
| + | * Click on the <span style="color:#3366ff;">'''Rec '''</span>button and select '''Original Average Reference''' in the dropdown menu to display all 65 electrodes using the average reference. | ||
| − | + | '''Step 2: Examine the data''' | |
| − | + | * Scan through the data to find the triggers for leftward horizontal eye movements (trigger 1). Double-click on an eye movement to view its topography. The figure shows the topography of a leftward eye movement. | |
| + | [[Image:Artifact Correction (12).gif]] | ||
| − | + | Note that the signal is always positive (red on the map) in the direction of the movement. | |
| + | (Note that older versions of calibration.foc contain three trigger codes: 1=horizontal, 2=vertical, 3=blink. The newer version contains five codes: 1=left, 2=right, 3=up, 4=down, 5=blink.) | ||
| + | * Look at maps of the vertical eye movements and the blinks. | ||
| − | |||
| − | + | '''Step 3: Pattern search and average: leftward eye movements''' | |
| − | + | * Find the first horizontal (leftward) eye movement and right click just before the onset of the movement. Select ''‘Default Block’ ''from the drop-down menu. The marked range should look something like this: | |
| − | + | [[Image:Artifact Correction (13).gif]] | |
| − | < | + | * Press the <span style="color:#3366ff;">'''SAW '''</span>(Search, Average, Write) button to start pattern search. For each segment that matches to the pattern, BESA Research will stop and ask for your confirmation. |
| + | * Accept only patterns for the onset of the eye movement (see the comments below). Reject patterns that are contaminated with blinks. Note the counter in the status bar indicating the number of averages. With this file, all 20 leftward eye movements can be included in the average. After 20 matches, pattern search may start matching up to the offset of the rightward eye movements. At this point you can press <span style="color:#3366ff;">'''Cancel'''</span> or the <span style="color:#3366ff;">'''Escape'''</span> key to terminate averaging. | ||
| + | * Save the averages in the suggested file, <span style="color:#ff9c00;">'''calibration.fsg'''</span>. Specify the label ''‘left’'' for the stored segment. | ||
| + | * Note that the onsets of leftward eye movements are now marked with the tag number 1. | ||
| − | |||
| + | '''Step 4: Other eye movements''' | ||
| − | <span style="color:#3366ff;"><span | + | * Repeat the above procedure for the rightward eye movements. First select buffer 2 for averaging. This will assign tag number 2 to these eye movements. |
| + | * Mark the first rightward eye movement as before. Press the <span style="color:#3366ff;">'''SAW'''</span> button. Again, 20 averages can be obtained. Label the average ''‘right’.'' | ||
| + | * Select buffer 3 for averaging. Repeat for the upward eye movements. 20 averages can be obtained. Use the label ''‘up’''. | ||
| + | * Select buffer 4 for averaging. Repeat for the downward eye movements. Note that the first downward eye movement is preceded by an upward saccade. Use the second movement as a template and reject the first movement from averaging. You will also need to reject the return movements from upward saccades. 19 averages will be obtained. Use the label ''‘down’''. Note that in this file these movements have a blink-like waveform at the onset. This may be due to eyelid movements. | ||
| + | * Select buffer 5 for averaging. Mark an eyeblink using a right click just before the onset of the blink. More than 20 averages can be obtained, because there are several blinks at the beginning of the data block. Make sure you do not include blinks that overlap with the eye movements. Use the label ''‘blink’''. Some subjects’ blinks are unnatural in the calibration file – see '''Comments on the Procedure''', below. | ||
| + | * Note that the different eye movements are marked with tags 1-5. If you need to repeat the averages (e.g. with different filter settings, a different time range), you do not need to repeat pattern search – you can use the search for tagged events (<span style="color:#3366ff;">'''Search/Tagged Events'''</span>) and switch off the Query option. Once the tags are defined, a repeat average can be done very quickly. The tags we generated are saved in the file <span style="color:#ff9c00;">'''calibration_pattern_search_result.evt'''</span>. These tags can be loaded using <span style="color:#3366ff;">'''ERP/Open Event File'''</span>… | ||
| − | |||
| − | + | '''Step 5: Examine the averages''' | |
| − | + | ||
| − | + | ||
| + | * Open the file, <span style="color:#ff9c00;">'''calibration.fsg'''</span>, you have just generated. Alternatively, open <span style="color:#ff9c00;">'''calibration_average.fsg'''</span>, the average file we generated. | ||
| + | * Turn off filters and select pre-stimulus baseline (<span style="color:#3366ff;">'''Filters/Use Prestimulus Baseline'''</span>). Set the display width to 6 s. | ||
| + | * Look at the maps of the five different segments. | ||
| − | |||
| − | + | '''Step 6: Define artifact topographies from the averaged data'''* We will now define artifact topographies from the averaged data: | |
| − | < | + | * Drag the cursor over the whole width of the horizontal eye movements (first two segments). |
| + | * Select <span style="color:#3366ff;">'''Artifact/Select'''</span>. In the resulting dialog, check the <span style="color:#3366ff;">'''HEOG'''</span> radio button. | ||
| − | |||
| − | + | [[Image:Artifact Correction (14).gif]] [[Image:Artifact Correction (15).gif]] | |
| − | |||
| − | < | + | * Note a) in the dropdown list to the right of the radio button, the variance explained by the first PCA component is usually in the high 90’s for averaged eye movements. This means: a single spatial component describes the eye movement extremely well! |
| + | * Note b) when the check box is clicked, the mapping window is opened automatically, displaying the topography of the spatial component. | ||
| + | * Note c) BESA Research allows to select up to three spatial components for each artifact category, using the dropdown list. This is useful for EKG correction in MEG data, where a single component is often not sufficient for correction. A use for eye correction is illustrated below. | ||
| + | * Press <span style="color:#3366ff;">'''OK'''</span> to close the dialog. Drag the cursor over the whole width of the vertical eye movements (3rd and 4th segment). | ||
| + | * Select <span style="color:#3366ff;">'''Artifact/Select.'''</span> In the resulting dialog, check the <span style="color:#3366ff;">'''HEOG'''</span> radio button. | ||
| − | |||
| − | + | [[Image:Artifact Correction (17).gif]] [[Image:Artifact Correction (16).gif]] | |
| − | |||
| − | + | * Drag the cursor over the width of the blink segment (5th segment). | |
| + | * Select <span style="color:#3366ff;">'''Artifact/Select'''</span>. In the resulting dialog, check the <span style="color:#3366ff;">'''BLINK'''</span> radio button. | ||
| − | + | [[Image:Artifact Correction (19).gif]] [[Image:Artifact Correction (18).gif]] | |
| − | < | + | * Press <span style="color:#3366ff;">'''OK '''</span>to close the dialog |
| + | * Note the appearance of the three artifact display channels at the bottom of the screen. Each channel images the activity of its eye movement type. Thus, you will see the horizontal eye movements on the right, the vertical movements in the middle, and the blink on the left. Especially during vertical eye movements, you will often see some blink activity – it seems that the eyelid is moving at the onset of the vertical movements (see figure below). | ||
| + | * Toggle artifact correction on and off using <span style="color:#3366ff;">'''Artifact/Correct'''</span> or the shortcut <span style="color:#3366ff;">'''ctrl-E'''</span>. To view the artifact correction signals more easily, set the EEG amplitude scaling to 200 µV, and the artifact correction signal scaling to 500 µV. Display the first quarter of the signals. Then the lower part of the display will look something like this: | ||
| − | |||
| − | < | + | <!-- [[Image:Artifact Correction (20).gif|600px]] --> |
| + | [[Image:Artifact Correction (20) new.png|800px]] | ||
| − | + | Note that correction is not perfect! In particular, some artifact signal will remain on electrodes near the eyes, e.g. E5, E14. In practice, this "imperfection" is not critical: most saccades are smaller than 15°, and blinks are usually corrected very well. | |
| − | < | + | * Save the artifact topographies in <span style="color:#ff9c00;">'''calibration.atf'''</span> (<span style="color:#3366ff;">'''Artifact/Save'''</span>). |
| − | |||
| − | + | '''Step 7: Generating correction coefficients using three orthogonal spatial components''' | |
| − | < | + | * Drag the mouse to highlight all five segments of the data. |
| + | * Select <span style="color:#3366ff;">'''Artifact/Select'''</span>. In the resulting dialog, un-check the '''check''' boxes to suppress the coefficients that were defined above. | ||
| + | * Check the<span style="color:#3366ff;">''' HEOG'''</span> radio button again. When asked whether to overwrite the existing coefficients, click the <span style="color:#3366ff;">'''Yes'''</span> button. As a result of deactivation and overwriting, the previously defined coefficients are removed, and new coefficients are allocated. | ||
| + | * Note that in the dropdown list, the first component explains about 66.5% of the data. Click on the arrow to the right of the list to show that the second component explains 29.9%, and the third component 3.1% of the data. Select the third component. This means that three components will be used for correction. | ||
| + | * Press <span style="color:#3366ff;">'''OK'''</span> to close the dialog. | ||
| − | |||
| − | + | [[Image:Artifact Correction (21).gif]] | |
| − | |||
| − | < | + | * Note that the '''HEOG''' artifact channel now displays three waveforms in different colors. These represent the three waveforms that are subtracted from the data to remove the artifacts. |
| + | * Save the artifact topographies in <span style="color:#ff9c00;">'''calibration_orthog.atf'''</span> (<span style="color:#3366ff;">'''Artifact/Save'''</span>). | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| + | '''Step 8: Test correction on the calibration data''' | ||
| − | * <span style="color:# | + | * Switch back to the raw data file, <span style="color:#ff9c00;">'''calibration.foc'''</span>. Select <span style="color:#3366ff;">'''Artifact/Load'''</span> and select the file <span style="color:#ff9c00;">'''calibration.atf'''</span>. <span style="color:#3366ff;">'''Artifact/Correct'''</span>. As with the averaged data, correction is quite good, but not perfect on electrodes near the eyes. |
| − | * <span style="color:#3366ff;"><span style="color:# | + | * Load <span style="color:#ff9c00;">'''calibration_orthog.atf'''</span> for comparison. Correction will be equivalent. |
| + | * What are the differences between these two approaches? The first method has the advantage that the artifact waveforms display the estimated activity of each type of eye movement: horizontal, vertical and blink. The second method combines these in a fairly arbitrary way, because the spatial topographies are orthogonal. The second method is simpler to apply. | ||
| + | * Close the raw data file (<span style="color:#3366ff;">'''File/Close'''</span>). Open it again. Note that the data are corrected, and the artifact waveforms are displayed. This is because <span style="color:#ff9c00;">'''calibration_orthog.atf'''</span> has been reloaded. Open the ''Load''<span style="color:#3366ff;">''' '''</span>''Coordinate Files'' dialog ('''''File/Digitized Coordinates/Head Surface Points and Sensors/Load Coordinate''''' '''''Files''''' or use the shortcut <span style="color:#3366ff;">'''ctrl-L'''</span>). At the bottom of the dialog, note that the file <span style="color:#ff9c00;">'''calibration_orthog.atf'''</span> has been selected in ''the "Artifact coefficients"'' section, because it has been associated with the data file in the BESA Research database. | ||
| + | * Select <span style="color:#ff9c00;">'''calibration.atf'''</span> and press <span style="color:#3366ff;">'''OK'''</span> to associate this file with the data, and press <span style="color:#3366ff;">'''OK'''</span>. | ||
| − | + | '''Comments on the procedure''' | |
| − | + | * '''Unnatural eyeblinks''': In some subjects, the eyeblinks are unnatural, e.g. too long, eyes shut too tightly. In this case, we use data from a measurement data block to collect an average of eyeblinks. | |
| − | + | * '''Overlapping saccades and eyeblinks''': Some subjects blink a lot during the saccades. This makes it difficult to obtain a good estimate of the eye movement topographies, because the eyeblink topography dominates. Solutions to this involve a) trying to use time ranges where the saccade dominates to estimate its topography, or b) using the first three components of a PCA of the combined averaged eye movements. | |
| − | * | + | * '''Filter settings''': the 0.5 – 8 Hz filters improves the signal-to-noise ratio of the eye activity. This filter setting is only used for averaging an estimating the eye movement topographies. For artifact correction, any filter setting can be used. |
| − | + | * '''Averaging to the onset of the eye movements:''' Pattern search often finds a match to the offset of an eye movement (e.g. searching for a leftward movement matches up to the return movements from the right saccade). In many cases, it would probably do no harm to the correction to include these segments in the average. However, we often find that the return movements are contaminated by blinks, so we usually exclude them from the averages. | |
| − | * | + | * '''Pre-stimulus baseline:''' The averages are generated so that the pre-stimulus baseline is before the eye movement or blink onset. When the pre-stimulus baseline option is selected on the average, we can obtain the topography of the eye movement relative to this baseline, i.e. relative to the fixation point. This is important for best correction results. Using a forward low cutoff filter ensures that the topography of the eye movement does not bleed back into the baseline. |
| − | * | + | * '''Time range''': Some subjects keep their eyes on the target for a shorter time. We generally adapt the time range for pattern search so that the end of the time range is in the middle of the target and not after movement offset. This avoids including blinks that may overlap with the offset of the eye movement. |
| − | + | [[Category:Research Manual]] | |
| − | + | {{BESAManualNav}} | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
Latest revision as of 14:03, 4 May 2021
| Module information | |
| Modules | BESA Research Basic or higher |
| Version | 6.1 or higher |
Contents
Introduction
BESA Research includes a sophisticated spatial filtering method to detect and correct artifacts. The method can be applied to any kind of artifact including EOG and EKG artifacts. Spatial topographies describing the artifacts may be defined manually or by allowing the program to scan the data automatically. Manual definition can be accomplished by marking a time range containing the artifact or a buffer with the averaged artifact. In particular, EKG artifacts should be averaged before defining the spatial topography. Once defined, the corrected data waveforms can be displayed, together with additional channels below the current montage showing the time course of the artifacts. Spatial artifact topographies can be saved to a file (*.atf), and may be recalled for later use with the same data or other data from the same subject.
Automatic artifact correction is described in the chapter Automatic Correction below. For a tutorial on using calibration data for an accurate estimation of artifact coefficients, see help chapter "Artifact Correction / Tutorial".
How correction works
Artifact correction is based on the spatial components method for correcting eye artifacts (Berg and Scherg 1994, see also Ille et al. 1997, 2002). Artifact and brain signal subspaces are described in terms of spatial topographies. The correction process consists of four steps:
- Define the topography for each type of artifact using automatic detection or manual definition
- Determine the brain signal topographies underlying the displayed EEG segment
- Reconstruct the artifact signal at each scalp electrode with a spatial filter taking into account artifact as well as brain signal subspace
- Subtract the reconstructed artifact signal from the original EEG segment
Available methods for correction
BESA Research allows artifact correction to be done in different ways. The method you choose depends on how you want to analyze your data. The methods vary in how the data is distorted by correction, and how the brain activity is modeled for the estimation of eye activity:
- Artifact-free review of the ongoing EEG. Use the PCA method (Ille et al. 2002). This is optimized to contrast the artifacts against the predominant rhythmic EEG activities.
- Analysis of averaged data waveforms (ERPs). Use the Surrogate method (Berg and Scherg 1994). Brain activity is modeled by a fixed dipole model (the "surrogate model"). Data waveforms will be distorted least if the Surrogate model is close to the dipole model of the averaged data.
- Source analysis. Use the Optimizing method (Berg and Scherg 1994). This is available in two variants:
- Develop dipole models in the presence of spatial components. This is the preferred method. Create separate averages of the uncorrected conditions and of the artifact events, e.g. blinks. All averages can be concatenated into one binary segment file (*.fsg). The averaged artifacts should then be used to define spatial components during source analysis. Thus, the time course of the artifacts and of the event-related source activity can be separated and observed in the source analysis window.
- Develop dipole models on the pre-corrected averages. Using subspace-projection (SSP), the artifact dimensions are compensated for in source models. This will lead to the same dipole models as above, but a) the time course of artifact activity is not available, and b) residual variance is lower because variance from dimensions in data space occupied by the artifact spatial components are removed from the data by SSP. It does not matter which correction method was used with BESA Research (PCA, Surrogate, or SSP) – the results of source analysis are the same.
- No artifact correction. If the data contain only a few artifacts, use artifact rejection rather than correction. This avoids the distortion resulting from correction.
Artifact correction in BESA Research distorts the data less than traditional eye artifact correction
Artifact correction can distort the data topographies and reduce or augment activities at some scalp regions, e.g. frontal scalp after eye movement correction. Therefore, maps should be interpreted with caution.
The advantage of the methods used in BESA Research over traditional approaches is that overlapping contributions of the artifacts and the EEG are separated as much as possible. In traditional approaches, a proportion of the signal on an EOG channel is always subtracted from each EEG signal. Thus, in contrast to traditional methods, the topography of the EEG is less distorted by correction based on spatial components, even if the correlation between artifact and brain activity is high.
Source Analysis takes distortion due to artifact correction into account
However much the signals may be distorted by artifact correction, the distortion can be taken into account in Source Analysis. This is accomplished using signal subspace projection (SSP, cf. Uusitalo et al. 1997), which is equivalent to the "optimizing" correction method described in Berg and Scherg (1994).
SSP is switched on in Source Analysis:
- if data were averaged with artifact correction switched on, and the averaged data were saved in BESA Research binary format (*.fsg).
- if artifact correction is switched on for any other type of file. For instance, you may have averaged a file to ASCII format. Read in the file, and load the artifact coefficients. Make sure correction is switched on.
The PCA and Surrogate methods distort less than traditional EOG subtraction methods, but they do not eliminate distortion! If you do not use the first variant of the Optimizing method in source analysis, you should make sure that subspace projection (SSP) is switched on in the source analysis module. This is indicated by the text "ART" at the right of the status bar in the source analysis module. Visual feedback that SSP is switched on is supplied in the Source Module by the text ART at the right of the Status Bar:
References
- Berg, P., and Scherg, M. A multiple source approach to the correction of eye artifacts, Electroenceph. clin. Neurophysiol., 1994, 90: 229-241.
- Ille N., Berg P., and Scherg M. A Spatial Components Method for Continuous Artifact Correction in EEG and MEG. Biomed. Tech., 1997, 42 (suppl. 1): 80-83.
- Ille, N., Berg, P., Scherg, M. Artifact correction of the ongoing EEG using spatial filters based on artifact and brain signal topographies. J. Clin. Neurophysiol. 2002, 19: 113-124.
- Uusitalo, M.A., Ilmoniemi, R.J. Signal-space projection method for separating MEG or EEG into components. Med. Biol. Eng. Comput., 1997, 35: 135-140.
Artifact Correction Menu
In the Review Window use the Artifact menu (Alt+A) to:
- assign EOG or ECG artifact topographies automatically
- assign artifacts manually
- display the reconstructed time course of the artifacts below the EEG
- display the EEG with the artifacts removed
- correct artifacts caused by simultaneous fMRI recording
- delete assigned artifacts
- select which artifact topographies to use
- display maps of the topography defining each artifact
- set options for modelling the brain signal subspace
- load a previously defined set of artifact topographies
- save the current set of artifact topographies to a file
Automatic Artifact Correction
BESA Research provides an automatic artifact correction method that enables users to quickly obtain artifact-cleared data. For more details see the main article on Automatic Artifact Correction
Using Defined Topographies for Correction
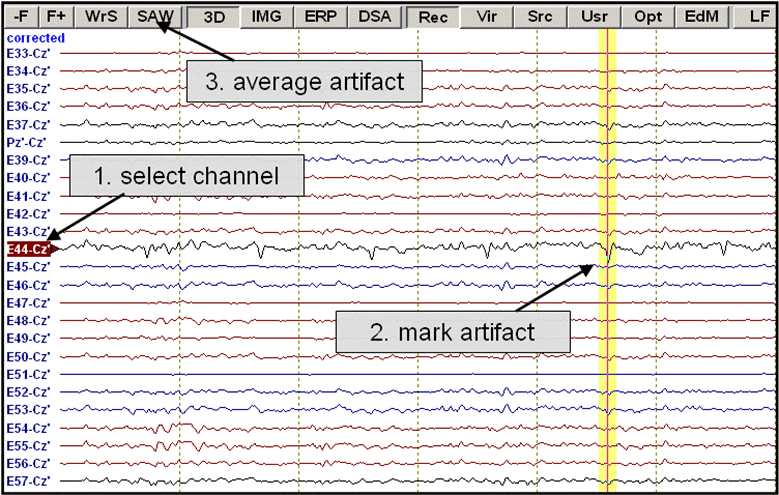
It is often desirable to use real data topographies for artifact correction, since they reflect the individual variability, e.g. in eye artifacts. The easiest way to assign an individual topography is to mark a data block which contains an artifact, and define the topography via the right click popup menu. The proceeding is as follows:
- Mark the artifact in the display window using the left mouse drag:
- Select the menu item Artifact / Select. Alternatively, right click and select Define Topography from the popup menu which appears:
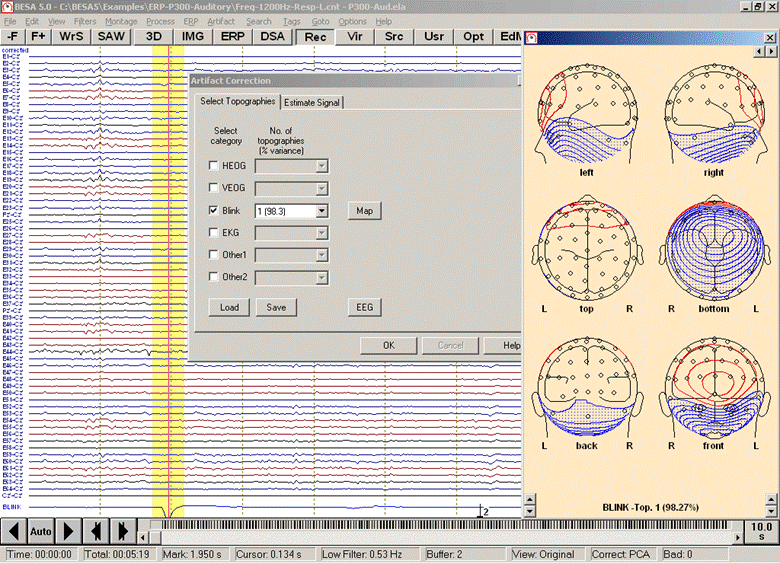
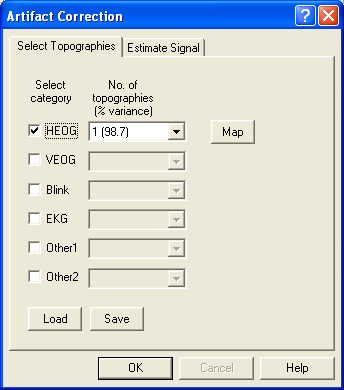
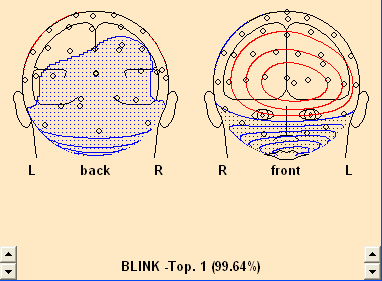
- The Select Topographies dialog box appears. There is a list of artifact types, for which topographies can be assigned. To assign a blink artifact, click into the check box in the third row. The topography is activated, and a map of the artifact topography is shown automatically:
EEG data after a topography has been assigned to the category "Blink". A map of the topography is shown at the right. An additional channel for the blink artifact is introduced at the bottom, and data are corrected automatically.
Note that the data are automatically corrected, and an additional virtual artifact channel appears at the bottom. To switch correction off again, use the menu item Artifact / Correct which toggles correction.
The artifact topography can be estimated with higher accuracy if a number of artifact epochs are averaged into a data buffer. This is especially recommended for correcting EKG artifacts, since the share of the overall topography which is held by the brain activity is larger for EKG artifacts. To average a number of artifacts:
- Select the channel where the artifact shows most prominently by a left click on the channel label.
- Mark the artifact in the data display.
- Press the SAW button at the top of the display window:
Averaging over an artifact
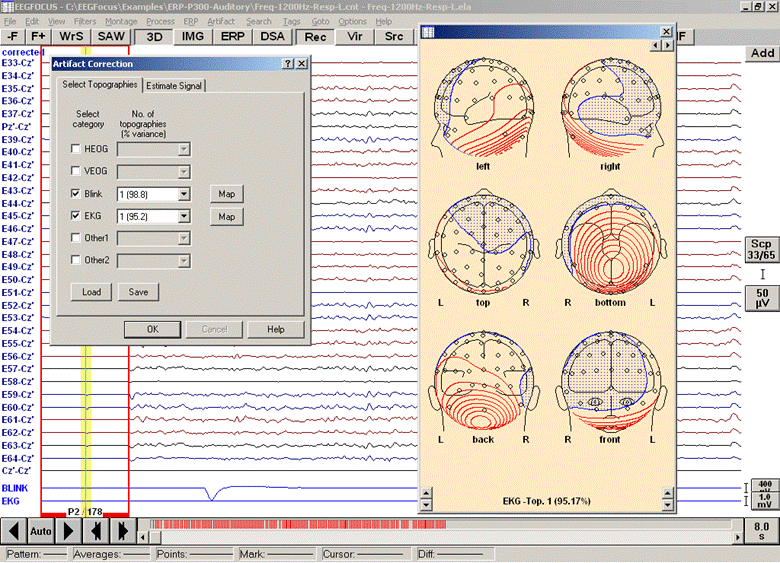
This starts an automatic average of all matching patterns in the data file. You may stop the search after a number of events were found (progress is shown in the status bar; stop averaging with the ESC key). When asked for a filename to save the pattern, press Cancel. The averaged data buffer containing the artifact is shown. Now use the averaged artifact to define the artifact topography as described above for the blink artifact. When selecting the artifact category, choose EKG:
Defining a topography for an EKG artifact from an averaged EKG pattern
For the correction of EKG artifacts, it is recommended to specify a (bipolar) polygraphic EKG channel. If only scalp channels are available, the correction can work reasonably only if more than 30 channels are available and if inferior electrodes are included.
Some artifacts can contain more than one spatial component (e.g. EKG artifacts). It is possible to select more than one component for correction.
More information on artifact correction is given in the help chapter "Artifact Correction / Tutorial".
fMRI artifact removal
(requires Besa Research 7.0 or higher)
Introduction
Currently data collected during simultaneous EEG-fMRI is not intended to be used in any diagnostic procedures. Following these guidelines, fMRI artifact removal implemented in BESA Research may also not be used for any diagnostic procedures This tool was developed only for research purposes.
Please use only EEG hardware that is certified for MRI!
When EEG data is recorded during an fMRI session there are plenty problems to solve to be able to analyze the real brain signal The first one is related to fast switching of MRI gradient coils which is a part of volume acquisition in the Echo-Planar sequence used for fMRI imaging This switching introduces variable magnetic fields that induce variable electrical fields in the EEG recording system (electrodes and wires namely), as described by Maxwell’s second law The induced artifact is typically very large in amplitude (typically 16 mV in a 3.0 T MR scanner). Also, the frequency range of the artifact starts in the frequency band of the EEG signal The artifact frequency and its harmonics can be easily assessed using the following equation (Rusiniak et al, 2013b):
where fx is the harmonic frequency, x is the harmonic number, NS represents the number of slices, and TR stands for Repetition Time of fMRI volume acquisition. From a variety of methods, the most relevant successful and best known are those based on moving average subtraction as firstly introduces by Allen and colleagues (2000). These methods work only when the following assumptions are true:
- Each artifact occurrence is identical: Each fMRI volume acquisition is identical (gradient switchingdoes not alter between volumes) so in general all artifacts are identical However, if the subject moves during the acquisition the artifact changes because electrode positions in relation to the local magnetic field have changed.
- EEG signal is stationary: In principle this assumption is true for the EEG signal. The signal stationaritycan be affected by the paradigm, therefore jitter between triggers is strongly advised.
- Each start points of artifacts have to be precisely indicated: This can be achieved either by hardwareclock synchronization between EEG system and MRI machine or by a very high sampling rate (>10 kHz). All modern EEG hardware units designed for simultaneous EEG-fMRI registration are capable of clock synchronization.
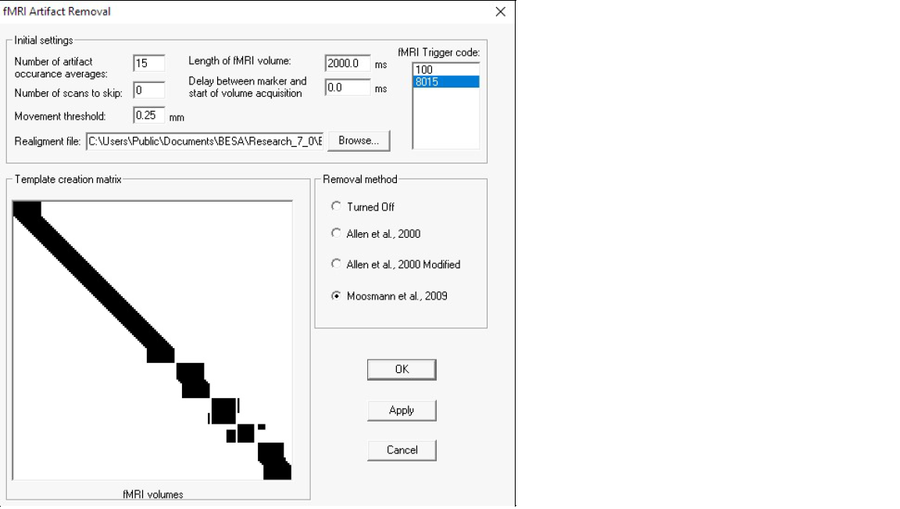
Parameters
- fMRI Trigger code The trigger code will be automatically detected in most cases If not please adjust this value to the correct code of trigger related to fMRI acquisition.
- Number of scans to average (k) Definition of how many surrounding artifact occurrences will be used for template creation (the larger the number, the more stable the artifact template will be, but in a dataset where there is a lot of subject movement the template will be far from good).
- Number of scans to skip This parameter indicates how many volumes from the beginning of the fMRI recording have to be skipped This setting is rarely used Use it only when scans that are not part of the actual fMRI sequence (i.e. dummy scans) have trigger events assigned that are visible in the EEG recording.
- Delay between marker and start of volume acquisition In the situation that volume acquisition starts before or after trigger occurrence this value can be used to adjust filtration.
- Length of fMRI volume (t) This value has to be equal to the fMRI sequence parameter – repetition time (TR) When a new file is opened this value is automatically computed assuming continuous EEG recording (fMRI volumes are recording without any gap).
- Movement threshold – This setting is only applicable for the last two filtration methods It indicates the minimum acceptable value of movement during the fMRI session.
- Realignment file A file that is the outcome of the step named “realignment” during fMRI data postprocessing BESA Research accepts files from SPM, BrainVoyager, AFNI, FSL The number of volumes in the realignment file has to match the number of triggers in the EEG data, and only then the last two removal methods will be available In rare cases use Number of scans to skip to adjust this value to match number of triggers between EEG and fMRI realignment file.
- Removal method In BESA, three methods are available (see below for more details) The last two methods are available only if a correctly configured realignment file is provided.
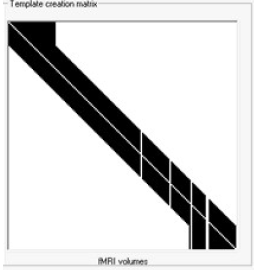
- Template creation matrix Visualization matrix that illustrates how the filtration is performed Check Filtration Methods description below for further details.
Filtration Methods
Allen et al, 2000
This method is a starting point for the two other methods as well In all methods a moving average artifact subtraction is used and the difference between methods can be noted in the template creation matrix To obtain artifact clean data (C) at epoch n the artifact template (B) has to be subtracted from artifact contaminated data (A) The mathematical principle for creating an artifact template at n-th epoch (Bn) is:
, where m is the number of fMRI volumes recorded, and k is the number of artifact volumes used for template creation (averaging).
After the artifact template for every epoch is created the final subtraction can be performed:
- Cn = A n - Bn
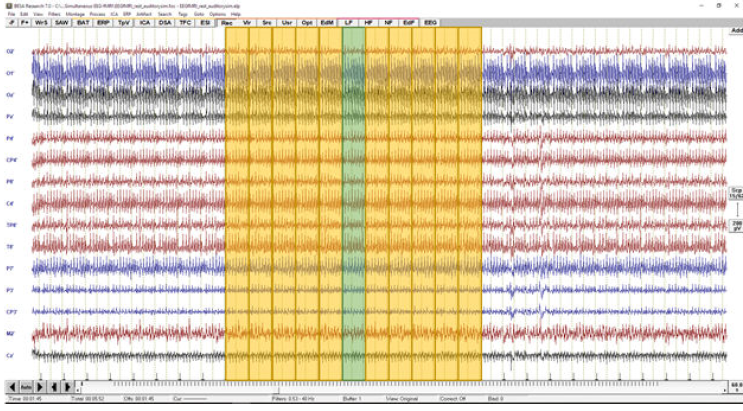
This procedure can be explained visually:
To clean the epoch indicated by the green shaded box the surrounding epochs have to be averaged first (here the averaging constant is k=10) After the template is created, it is subtracted from the recorded data at each channel independently to obtain artifact free data Next, this procedure is repeated for every epoch related to fMRI acquisition as indicated in the template creation matrix:
Note how the template changes when the number of artifact occurrence used for averaging is increased to k=30:
Allen et al, 2000 Modified
This method is available only if a correctly configured realignment file is provided Also, the changes in filtration will be visible if there is significant movement in the data (a recommended value for the movement threshold is 0.3 mm) In the modified Allen method, the epochs where subject’s movement is exceeding the threshold are ignored during artifact template creation, as shown in the template creation matrix below Note the vertical white lines – they represent the moments were movement was higher than threshold The data will be filtered but less artifact occurrences will be used for template creation close to a movement area.
Moosmann et al, 2009
As with the modified Allen method, this method (Moosmann et al, 2009) is also available only if a correctly configured realignment file is provided. The adjustment of the template creation matrix is much more advanced and can be explained by the following equation
, where all parameters are defined as above and z means an epoch number where movement exceeds the threshold This operation results in the following template creation matrix:
Note that this operation results in a different shape of matrix Importantly there are also white vertical lines This indicates that there was more than one epoch contaminated with movement In such situations, the matrix is transformed to adjust to fit to the largest movements, and smaller movements are treated as in Allen et al, 2000 Modified method.
Good Practice
Please keep in mind that simultaneous EEG-fMRI recording is a difficult yet powerful technique Following these rules could help one to perform a successful experiment:
- For an ERP experiment remember to design it with jitter between trials (i.e. random value of ±200 ms) Also, applying a pure EEG or pure fMRI paradigm will probably have the effect that one of the modalities will not show satisfying results Proper paradigm preparation is essential for success Some further guidelines can be found here:(Rusiniak et al, 2013a)
- Inform your subject how important it is not to move during the acquisition.
- Keep electrode to skin impedance as low as possible.
- The EEG-fMRI recording session should be long enough to allow for proper artifact creation Usually an experiment should last at least 6 minutes.
- At the same time try to limit time of experiment to a minimum and preferably perform EEG-fMRI registration before other sequences to limit movement due to inconvenient supine position.
- Place subject about 4 cm further in caudal direction to reduce artifacts The MRI laser crosshair should point not to the Nasion position but to the middle of the forehead (Mullinger et al, 2011)
- Especially for the first few registrations repeat the experiment outside of the bore to compare results
- Remember that conditions in MRI are far from standard EEG recording The long supine position, electrode pressure on back of the head, huge noise and stress can affect the EEG!
Balistocardiogram and other artifacts
After removing the fMRI gradient artifact many different steps can be performed (Abreu et al, 2018) For ECG induced artifacts (balistocardiogram) we recommend artifact correction using defined topographies for correction (usually with more than 3 components to filter).
References
- Abreu, R, Leal, A, Figueiredo, P, 2018 EEG-Informed fMRI A Review of Data Analysis Methods Front Hum Neurosci 12, 29 https://doi.org/10.3389/fnhum.2018.00029
- Allen, P.J., Josephs, O, Turner, R, 2000 A Method for Removing Imaging Artifact from Continuous EEG Recorded during Functional MRI. NeuroImage 12, 230–239 https://doi.org/10.1006/nimg.2000.0599
- Moosmann, M, Schönfelder, V.H., Specht, K, Scheeringa, R, Nordby, H, Hugdahl, K, 2009. Realignment parameter-informed artefact correction for simultaneous EEG–fMRI recordings. NeuroImage 45, 1144–1150 https://doi.org/10.1016/j.neuroimage.2009.01.024
- Mullinger, K.J., Yan, W.X., Bowtell, R, 2011. Reducing the gradient artefact in simultaneous EEG-fMRI by adjusting the subject’s axial position. NeuroImage 54, 1942–1950, https://doi.org/10.1016/j.neuroimage.2010.09.079
- Rusiniak, M, Lewandowska, M, Wolak, T, Pluta, A, Milner, R, Ganc, M, Włodarczyk, A, Senderski, A, Śliwa, L, Skarżyński, H, 2013a. A modified oddball paradigm for investigation of neural correlates of attention a simultaneous ERP–fMRI study Magn Reson Mater Phys Biol Med 26, 511–526 https://doi.org/10.1007/s10334-013-0374-7
- Rusiniak, M, Wolak, T, Lewandowska, M, Cieśla, K, Skarzynski, H, 2013b. The relation between EPI sequence parameters and electroencephalographic data during simultaneus EEG-fMRI registration an initial report, in ESMRMB 2013 Congress, Book of Abstracts, Saturday Presented at the ESMRMB, Springer, Toulouse, p 661 https://doi.org/10.1007/s10334-013-0384-5
Artifacts Tutorial: Using Calibration Data to Generate Artifact Coefficients
In this example, we will work on calibration data containing systematic eye movements:
- Each type of eye movement (left, right, up, down, blink) will be averaged separately, using pattern search.
- Correction coefficients for horizontal and vertical eye movements and blinks will be generated.
- We will obtain feedback about the coefficients: how well do these coefficients correct the calibration data?
- Using the PCA in the source analysis module, we can demonstrate the presence of rider artifacts (small eyelid movements) overlapping the saccades.
The data example, Examples\Xtras\Eye Artifacts\calibration.foc, was collected using 64 channels referred to Cz. In the eye movement calibration procedure, the subject watched a fixation point on a video monitor. At intervals of 2 s, an arrow appeared on the screen. The subject was instructed to move his eyes in the direction of the arrow to a point (marked on the wall of the shielded cabin) approximately 15° away from the fixation point, and then back to the fixation point. The subject was also instructed to try not to blink during the eye movement, but was allowed to blink while watching the fixation point. There were 20 movements in each of the directions left, right, up, down. Following these movements, the arrows were replaced by an image of an eye. This was a cue to blink. 20 eyeblinks were requested.
In the data file, trigger 1 denotes the horizontal arrows, trigger 2 the vertical arrows, and trigger 3 the eyeblinks.
Step 1: Setting up the parameters
- Load the file, Examples\Xtras\Eye Artifacts\calibration.foc.
- Set the Default Block Epoch to the range -100 ms to +400 ms (Edit/Default Block Epoch).
- Select pattern search (Search/Pattern), and make sure that the Query option is switched on (Search/Query).
- Select buffer 1 for averaging (Tags/Pattern 1).
- Set the filters to 0.5 Hz high pass, 6 db/octave, forward, and 8 Hz low pass, 12 db/octave, zero phase (Filters/Edit Filter Settings).
- Click on the Rec button and select Original Average Reference in the dropdown menu to display all 65 electrodes using the average reference.
Step 2: Examine the data
- Scan through the data to find the triggers for leftward horizontal eye movements (trigger 1). Double-click on an eye movement to view its topography. The figure shows the topography of a leftward eye movement.
Note that the signal is always positive (red on the map) in the direction of the movement. (Note that older versions of calibration.foc contain three trigger codes: 1=horizontal, 2=vertical, 3=blink. The newer version contains five codes: 1=left, 2=right, 3=up, 4=down, 5=blink.)
- Look at maps of the vertical eye movements and the blinks.
Step 3: Pattern search and average: leftward eye movements
- Find the first horizontal (leftward) eye movement and right click just before the onset of the movement. Select ‘Default Block’ from the drop-down menu. The marked range should look something like this:
- Press the SAW (Search, Average, Write) button to start pattern search. For each segment that matches to the pattern, BESA Research will stop and ask for your confirmation.
- Accept only patterns for the onset of the eye movement (see the comments below). Reject patterns that are contaminated with blinks. Note the counter in the status bar indicating the number of averages. With this file, all 20 leftward eye movements can be included in the average. After 20 matches, pattern search may start matching up to the offset of the rightward eye movements. At this point you can press Cancel or the Escape key to terminate averaging.
- Save the averages in the suggested file, calibration.fsg. Specify the label ‘left’ for the stored segment.
- Note that the onsets of leftward eye movements are now marked with the tag number 1.
Step 4: Other eye movements
- Repeat the above procedure for the rightward eye movements. First select buffer 2 for averaging. This will assign tag number 2 to these eye movements.
- Mark the first rightward eye movement as before. Press the SAW button. Again, 20 averages can be obtained. Label the average ‘right’.
- Select buffer 3 for averaging. Repeat for the upward eye movements. 20 averages can be obtained. Use the label ‘up’.
- Select buffer 4 for averaging. Repeat for the downward eye movements. Note that the first downward eye movement is preceded by an upward saccade. Use the second movement as a template and reject the first movement from averaging. You will also need to reject the return movements from upward saccades. 19 averages will be obtained. Use the label ‘down’. Note that in this file these movements have a blink-like waveform at the onset. This may be due to eyelid movements.
- Select buffer 5 for averaging. Mark an eyeblink using a right click just before the onset of the blink. More than 20 averages can be obtained, because there are several blinks at the beginning of the data block. Make sure you do not include blinks that overlap with the eye movements. Use the label ‘blink’. Some subjects’ blinks are unnatural in the calibration file – see Comments on the Procedure, below.
- Note that the different eye movements are marked with tags 1-5. If you need to repeat the averages (e.g. with different filter settings, a different time range), you do not need to repeat pattern search – you can use the search for tagged events (Search/Tagged Events) and switch off the Query option. Once the tags are defined, a repeat average can be done very quickly. The tags we generated are saved in the file calibration_pattern_search_result.evt. These tags can be loaded using ERP/Open Event File…
Step 5: Examine the averages
- Open the file, calibration.fsg, you have just generated. Alternatively, open calibration_average.fsg, the average file we generated.
- Turn off filters and select pre-stimulus baseline (Filters/Use Prestimulus Baseline). Set the display width to 6 s.
- Look at the maps of the five different segments.
Step 6: Define artifact topographies from the averaged data* We will now define artifact topographies from the averaged data:
- Drag the cursor over the whole width of the horizontal eye movements (first two segments).
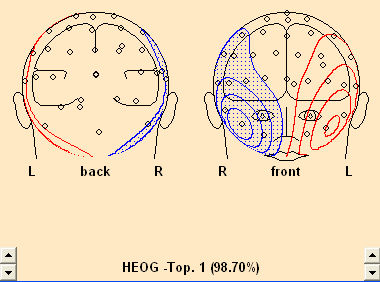
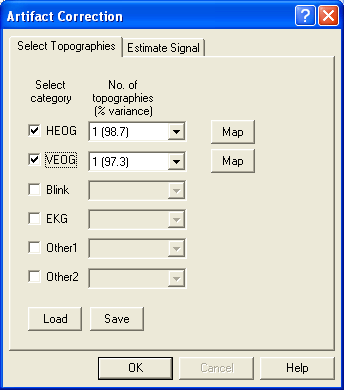
- Select Artifact/Select. In the resulting dialog, check the HEOG radio button.
- Note a) in the dropdown list to the right of the radio button, the variance explained by the first PCA component is usually in the high 90’s for averaged eye movements. This means: a single spatial component describes the eye movement extremely well!
- Note b) when the check box is clicked, the mapping window is opened automatically, displaying the topography of the spatial component.
- Note c) BESA Research allows to select up to three spatial components for each artifact category, using the dropdown list. This is useful for EKG correction in MEG data, where a single component is often not sufficient for correction. A use for eye correction is illustrated below.
- Press OK to close the dialog. Drag the cursor over the whole width of the vertical eye movements (3rd and 4th segment).
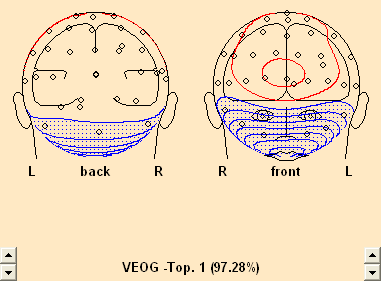
- Select Artifact/Select. In the resulting dialog, check the HEOG radio button.
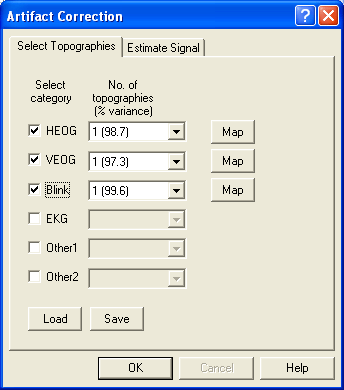
- Drag the cursor over the width of the blink segment (5th segment).
- Select Artifact/Select. In the resulting dialog, check the BLINK radio button.
- Press OK to close the dialog
- Note the appearance of the three artifact display channels at the bottom of the screen. Each channel images the activity of its eye movement type. Thus, you will see the horizontal eye movements on the right, the vertical movements in the middle, and the blink on the left. Especially during vertical eye movements, you will often see some blink activity – it seems that the eyelid is moving at the onset of the vertical movements (see figure below).
- Toggle artifact correction on and off using Artifact/Correct or the shortcut ctrl-E. To view the artifact correction signals more easily, set the EEG amplitude scaling to 200 µV, and the artifact correction signal scaling to 500 µV. Display the first quarter of the signals. Then the lower part of the display will look something like this:
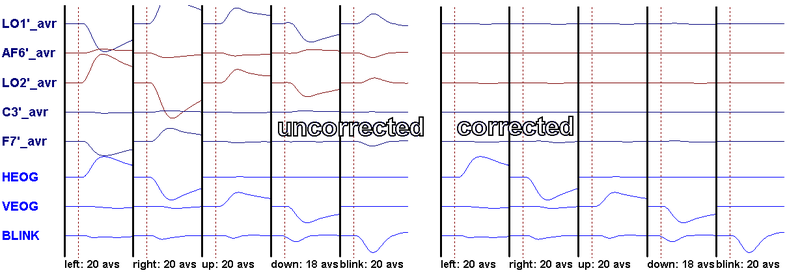
Note that correction is not perfect! In particular, some artifact signal will remain on electrodes near the eyes, e.g. E5, E14. In practice, this "imperfection" is not critical: most saccades are smaller than 15°, and blinks are usually corrected very well.
- Save the artifact topographies in calibration.atf (Artifact/Save).
Step 7: Generating correction coefficients using three orthogonal spatial components
- Drag the mouse to highlight all five segments of the data.
- Select Artifact/Select. In the resulting dialog, un-check the check boxes to suppress the coefficients that were defined above.
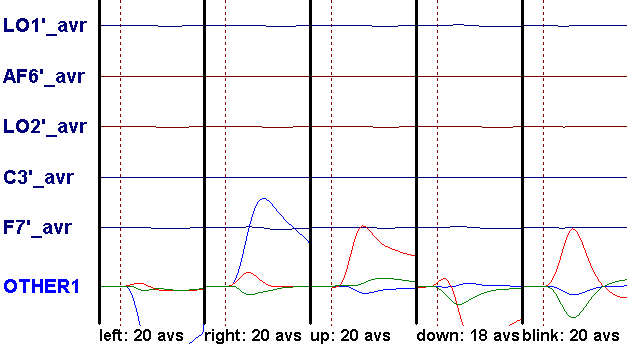
- Check the HEOG radio button again. When asked whether to overwrite the existing coefficients, click the Yes button. As a result of deactivation and overwriting, the previously defined coefficients are removed, and new coefficients are allocated.
- Note that in the dropdown list, the first component explains about 66.5% of the data. Click on the arrow to the right of the list to show that the second component explains 29.9%, and the third component 3.1% of the data. Select the third component. This means that three components will be used for correction.
- Press OK to close the dialog.
- Note that the HEOG artifact channel now displays three waveforms in different colors. These represent the three waveforms that are subtracted from the data to remove the artifacts.
- Save the artifact topographies in calibration_orthog.atf (Artifact/Save).
Step 8: Test correction on the calibration data
- Switch back to the raw data file, calibration.foc. Select Artifact/Load and select the file calibration.atf. Artifact/Correct. As with the averaged data, correction is quite good, but not perfect on electrodes near the eyes.
- Load calibration_orthog.atf for comparison. Correction will be equivalent.
- What are the differences between these two approaches? The first method has the advantage that the artifact waveforms display the estimated activity of each type of eye movement: horizontal, vertical and blink. The second method combines these in a fairly arbitrary way, because the spatial topographies are orthogonal. The second method is simpler to apply.
- Close the raw data file (File/Close). Open it again. Note that the data are corrected, and the artifact waveforms are displayed. This is because calibration_orthog.atf has been reloaded. Open the Load Coordinate Files dialog (File/Digitized Coordinates/Head Surface Points and Sensors/Load Coordinate Files or use the shortcut ctrl-L). At the bottom of the dialog, note that the file calibration_orthog.atf has been selected in the "Artifact coefficients" section, because it has been associated with the data file in the BESA Research database.
- Select calibration.atf and press OK to associate this file with the data, and press OK.
Comments on the procedure
- Unnatural eyeblinks: In some subjects, the eyeblinks are unnatural, e.g. too long, eyes shut too tightly. In this case, we use data from a measurement data block to collect an average of eyeblinks.
- Overlapping saccades and eyeblinks: Some subjects blink a lot during the saccades. This makes it difficult to obtain a good estimate of the eye movement topographies, because the eyeblink topography dominates. Solutions to this involve a) trying to use time ranges where the saccade dominates to estimate its topography, or b) using the first three components of a PCA of the combined averaged eye movements.
- Filter settings: the 0.5 – 8 Hz filters improves the signal-to-noise ratio of the eye activity. This filter setting is only used for averaging an estimating the eye movement topographies. For artifact correction, any filter setting can be used.
- Averaging to the onset of the eye movements: Pattern search often finds a match to the offset of an eye movement (e.g. searching for a leftward movement matches up to the return movements from the right saccade). In many cases, it would probably do no harm to the correction to include these segments in the average. However, we often find that the return movements are contaminated by blinks, so we usually exclude them from the averages.
- Pre-stimulus baseline: The averages are generated so that the pre-stimulus baseline is before the eye movement or blink onset. When the pre-stimulus baseline option is selected on the average, we can obtain the topography of the eye movement relative to this baseline, i.e. relative to the fixation point. This is important for best correction results. Using a forward low cutoff filter ensures that the topography of the eye movement does not bleed back into the baseline.
- Time range: Some subjects keep their eyes on the target for a shorter time. We generally adapt the time range for pattern search so that the end of the time range is in the middle of the target and not after movement offset. This avoids including blinks that may overlap with the offset of the eye movement.
| Review | |
|---|---|
| Source Analysis | |
| Integration with MRI and fMRI | |
| Source Coherence | |
| Export | |
| MATLAB Interface | |
| Special Topics | |