Difference between revisions of "Working With Additional Files"
(→ASCII Multiplexed format (*.mul)) |
|||
| (10 intermediate revisions by one other user not shown) | |||
| Line 2: | Line 2: | ||
|title = Module information | |title = Module information | ||
|module = BESA Research Basic or higher | |module = BESA Research Basic or higher | ||
| − | |version = 6.1 or higher | + | |version = BESA Research 6.1 or higher |
}} | }} | ||
| − | |||
| − | + | == Binary format (*.foc, *.fsg) == | |
| − | Select '''Binary High Resolution''' or '''Binary Compressed Format''' to output segments in binary BESA format. If the file already exists, the segment will be appended. Thus, it is possible to create a file combining several segments of interest in a compact form. BESA Research will only allow you to append segments if the number of channels and the sampling interval in source and target files are the same. In Binary Format all channels (scalp, intracranial, polygraphic, MEG) and file events in the selected time range are exported. '''Note:''' The channels are filtered according to the current filter settings. | + | Select '''Binary High Resolution''' or '''Binary Compressed Format''' to output segments in binary BESA format. If the file already exists, the segment will be appended. Thus, it is possible to create a file combining several segments of interest in a compact form. BESA Research will only allow you to append segments if the number of channels and the sampling interval in source and target files are the same. |
| + | |||
| + | In Binary Format all channels (scalp, intracranial, polygraphic, MEG) and file events in the selected time range are exported. '''Note:''' The channels are filtered according to the current filter settings. | ||
Select '''Binary High Resolution''' to retain the resolution of the processed data. This is the preferred binary format for small amplitude signals such as averages. Select '''Binary Compressed Format''' to store raw data using the original resolution or to obtain the space savings of compression (see ''File/Export and Append Data/Convert..'' above). This is the preferred binary format for raw data. | Select '''Binary High Resolution''' to retain the resolution of the processed data. This is the preferred binary format for small amplitude signals such as averages. Select '''Binary Compressed Format''' to store raw data using the original resolution or to obtain the space savings of compression (see ''File/Export and Append Data/Convert..'' above). This is the preferred binary format for raw data. | ||
| − | + | == ASCII vectorized format (*.avr) == | |
Select '''ASCII vectorized Format''' to output segments in BESA ASCII format, one channel (all time points) per line. | Select '''ASCII vectorized Format''' to output segments in BESA ASCII format, one channel (all time points) per line. | ||
| + | |||
'''The Format is as follows:''' | '''The Format is as follows:''' | ||
| Line 21: | Line 23: | ||
The first of two header lines contains the following data descriptors (6 descriptors, the values shown are only examples): | The first of two header lines contains the following data descriptors (6 descriptors, the values shown are only examples): | ||
| − | + | {| | |
| − | ''Npts= 200'' | + | | width="30%" |''Npts= 200'' || number of sampled points in each channel |
| − | + | |- | |
| − | ''TSB= -500'' | + | | ''TSB= -500'' || time sweep begin [ms]. Time of first data point relative to zero of epoch |
| − | + | |- | |
| − | ''DI= 5'' | + | | ''DI= 5'' || digitization or sampling interval [ms] |
| − | + | |- | |
| − | ''SB= 2'' | + | | ''SB= 2'' || scaling bins/microvolt in file = number corresponding to 1 microvolt |
| − | + | |- | |
| − | ''SC= 50'' | + | | ''SC= 50'' || scaling calibration, used for setting magnitude of display in BESA |
| − | + | |- | |
| − | ''Nchan= 27'' | + | | ''Nchan= 27'' || number of channels |
| − | + | |- | |
| − | ''SegmentName= 60dB'' | + | | ''SegmentName= 60dB'' || an optional label describing the data |
| + | |} | ||
The second line of the header contains a label for each channel, e.g. | The second line of the header contains a label for each channel, e.g. | ||
| Line 41: | Line 44: | ||
| − | Each of the subsequent ''Nchan'' lines of the file contains values for all ''Npts'' time points in floating point or scientific format. For more details about scalp electrodes, see chapter | + | Each of the subsequent ''Nchan'' lines of the file contains values for all ''Npts'' time points in floating point or scientific format. For more details about scalp electrodes, see chapter ''[[Electrodes_and_Surface_Locations#Electrode_Conventions|Working With Electrodes and Surface Locations/Electrodes/Electrode Conventions]]''. |
| − | A second (older) version of the format (written by BESA versions 1, 2 and 3) omits the '''Nchan=xx''' information in the first line, and there is no second header line. Labels must be defined elsewhere. See | + | A second (older) version of the format (written by BESA versions 1, 2 and 3) omits the '''<tt>Nchan=xx</tt>''' information in the first line, and there is no second header line. Labels must be defined elsewhere. See ''[[Working_With_Additional_Files#Channel_definition_file_conventions_and_formats|Channel definition file conventions and formats]]'' and ''[[Electrodes_and_Surface_Locations#Reading_MEG_files_in_ASCII_format|Reading MEG files in ASCII format]]''. In the older versions, only scalp channels were exported, and the data were average referenced. |
| + | == ASCII Multiplexed format (*.mul) == | ||
| − | + | Select '''ASCII Multiplexed Format''' to output segments one time point (all channels) per line. | |
| − | + | ||
| − | Select '''ASCII Multiplexed Format''' to output segments one time point (all channels) per line. | + | |
The '''ASCII Multiplexed Format '''is as follows: | The '''ASCII Multiplexed Format '''is as follows: | ||
| + | |||
The first of two header lines contains similar information to that of the ASCII vectorized file: | The first of two header lines contains similar information to that of the ASCII vectorized file: | ||
| + | <source lang="dos"> | ||
| + | TimePoints= 200 Channels= 27 BeginSweep[ms]= -500.00 SamplingInterval[ms]= 5.000 Bins/uV= 1.000 SegmentName=Condition1 | ||
| + | </source> | ||
| − | '' | + | Note that the item '''<tt>SegmentName</tt>''' is missing if no segment comment is specified when writing a segment to file. |
| − | + | If an epoch of a continuous EEG is exported in ASCII multiplexed format, the first header line contains the additional item '''<tt>Time</tt>''', which indicates the daytime of the first sample in the exported segment: | |
| − | + | <source lang="dos"> | |
| + | TimePoints= 200 Channels= 27 BeginSweep[ms]= 0.00 SamplingInterval[ms]= 5.000 Bins/uV= 1.000 Time=22:02:53 SegmentName=Segment1 | ||
| + | </source> | ||
| − | |||
The second line of the header contains labels for each channel, which may be either the original channel names, or the names of the channels of the current montage, e.g. | The second line of the header contains labels for each channel, which may be either the original channel names, or the names of the channels of the current montage, e.g. | ||
| − | + | <source lang="dos"> | |
| + | O1 Oz P3 T5 T3 C3 F7 F3 Fp1 Fz Cz Pz Fp2 F4 F8 C4 T4 T6 P4 Fpz O2 M2 M1 F10 F9 T10 T9 | ||
| + | </source> | ||
Each subsequent line contains values for all 'Channels' at one time point, in floating point or scientific format. Values are given for the current or the original montage, selected as described above. | Each subsequent line contains values for all 'Channels' at one time point, in floating point or scientific format. Values are given for the current or the original montage, selected as described above. | ||
| Line 87: | Line 96: | ||
* The final letter after the hyphen indicates L=left, M=middle, R=right. | * The final letter after the hyphen indicates L=left, M=middle, R=right. | ||
| − | + | == Channel definition file conventions and formats == | |
BESA Research can read 3 types of file to define channels. These are identified by different extensions: | BESA Research can read 3 types of file to define channels. These are identified by different extensions: | ||
| − | * | + | * <span style="color:#ff9c00;">'''<tt>*.ela</tt>'''</span>: channel definition files containing labels and, optionally, channel types (ASCII, 1 label / line) |
| − | + | * <span style="color:#ff9c00;">'''<tt>*.elp</tt>'''</span>: channel definition files containing coordinates and, optionally, channel types and labels (ASCII). This is the format of the file written by the ''Channel Configuration Dialog.'' | |
| − | * | + | * <span style="color:#ff9c00;">'''<tt>*.elb</tt>'''</span>: channel definition files stored by older versions of BESA Research (binary format). This format can still be read, but is no longer written by BESA Research. |
| Line 98: | Line 107: | ||
# For all data files with the extension <span style="color:#ff9c00;">'''.eeg'''</span>, <span style="color:#ff9c00;">'''.cnt'''</span> and <span style="color:#ff9c00;">'''.foc'''</span>, check in the additional database file in the data directory with the same basename as the data file and the extension <span style="color:#ff9c00;">'''.fst'''</span>, whether a channel file has been associated previously | # For all data files with the extension <span style="color:#ff9c00;">'''.eeg'''</span>, <span style="color:#ff9c00;">'''.cnt'''</span> and <span style="color:#ff9c00;">'''.foc'''</span>, check in the additional database file in the data directory with the same basename as the data file and the extension <span style="color:#ff9c00;">'''.fst'''</span>, whether a channel file has been associated previously | ||
| − | # Check in the | + | # Check in the '''db''' subdirectory whether a channel file has been associated previously |
# Check if labels are defined in the header of the data file | # Check if labels are defined in the header of the data file | ||
# Search for a corresponding binary channel definition file with the same basename as the data file in the data folder (<span style="color:#ff9c00;">'''xxxx.elb'''</span>) | # Search for a corresponding binary channel definition file with the same basename as the data file in the data folder (<span style="color:#ff9c00;">'''xxxx.elb'''</span>) | ||
| Line 109: | Line 118: | ||
| − | '''Channel Label Files (*.ela)''' | + | '''Channel Label Files (<span style="color:#ff9c00;">*.ela</span>)''' |
Files containing a list of channel labels are an alternative to editing electrode configurations. They can be edited using a standard text editor. Electrode label files (<span style="color:#ff9c00;">'''.ela'''</span>) require a sequence of lines corresponding to the sequence of channels in the data. Each line contains one label and an optional identifier. The format is ' [Identifier] {Label} ', ('Identifier' can be omitted if the electrode label defines the type of signal) | Files containing a list of channel labels are an alternative to editing electrode configurations. They can be edited using a standard text editor. Electrode label files (<span style="color:#ff9c00;">'''.ela'''</span>) require a sequence of lines corresponding to the sequence of channels in the data. Each line contains one label and an optional identifier. The format is ' [Identifier] {Label} ', ('Identifier' can be omitted if the electrode label defines the type of signal) | ||
| Line 115: | Line 124: | ||
'''Identifiers''' can be one of: | '''Identifiers''' can be one of: | ||
| − | EEG | + | * EEG - scalp electrode |
| − | + | * SCP - scalp electrode | |
| − | SCP | + | * POL - polygraphic channel |
| − | + | * PGR - polygraphic channel | |
| − | POL | + | * ICR - intracranial electrode |
| − | + | * MEG - MEG sensor | |
| − | PGR | + | * REF - reference electrode (this can only occur once, and must be the last item in the file) |
| − | + | ||
| − | ICR | + | |
| − | + | ||
| − | MEG | + | |
| − | + | ||
| − | REF | + | |
| − | + | ||
For example: | For example: | ||
| − | + | <blockquote style="background-color: lightgrey; border: solid thin grey;"> | |
| + | Fz ('''scalp''' electrode, coordinates assigned by default.ecd) | ||
| − | Cz | + | Cz ('''scalp '''electrode, coordinates assigned by default.ecd) |
...... | ...... | ||
| − | VEOG | + | VEOG (vertical EOG,''' Polygraphic''' type is assigned by default) |
| − | Exx | + | Exx (xx=01, 02.. electrode number, '''Polygraphic '''type is assigned by default) |
...... | ...... | ||
| Line 148: | Line 151: | ||
...... | ...... | ||
| − | POL XX | + | POL XX (identifier sets '''Polygraphic''' type -- an alternative to the "POL" prefix is "PGR") |
...... | ...... | ||
| − | ICR | + | ICR A01 (identifier sets '''Intracranial''' type to electrode A01 - do not use A1!) |
| − | ICR | + | ICR A02 (identifier sets''' Intracranial''' type to electrode A02 - do not use A2!) |
| − | ICR | + | ICR A03 (identifier sets''' Intracranial''' type to electrode A03) |
...... | ...... | ||
| − | MEG | + | MEG M01 (identifier sets '''MEG '''type to electrode M01 - do not use M1!) |
| − | MEG | + | MEG M02 (identifier sets '''MEG''' type to electrode M02 - do not use M2!) |
...... | ...... | ||
| − | REF Cz | + | REF Cz (the label is assigned to the electrode reference, no channel is associated with this entry. This must be the last line of the file!) |
| + | </blockquote> | ||
| − | '''Channel spherical coordinate files (*.elp)''' | + | '''Channel spherical coordinate files (<span style="color:#ff9c00;">*.elp</span>)''' |
| − | These files follow the same rules as the channel label files, with the addition of spherical coordinates (theta, phi) that follow the labels of EEG and MEG channels. Labels can also be omitted. In this case, BESA Research will assign labels according to the nearest coordinate defined in the <span style="color:#ff9c00;">'''default.ecd'''</span> file. To indicate that the coordinates have been assigned, the label will have a tick, e.g. Fz' instead of Fz. | + | These files follow the same rules as the channel label files, with the addition of spherical coordinates (theta, phi) that follow the labels of EEG and MEG channels. Labels can also be omitted. In this case, BESA Research will assign labels according to the nearest coordinate defined in the <span style="color:#ff9c00;">'''default.ecd'''</span> file. To indicate that the coordinates have been assigned, the label will have a tick, e.g. '''<span style="color:#3366ff;">Fz'</span>''' instead of '''Fz'''. |
| − | Channels of other types (polygraphic, intracranial) are defined exactly as in the channel label (<span style="color:#ff9c00;">''' | + | Channels of other types (polygraphic, intracranial) are defined exactly as in the channel label (<span style="color:#ff9c00;">'''*.ela'''</span>) file. |
| − | + | == cot (Head center) file == | |
| − | + | ||
'''Function''': to redefine the center of the head for the sphere used in dipole models. If the cot file has the same base name as the data file, it is read automatically by BESA Research. If the coordinates deviate by more than 1 mm from the previously defined head center, a window is opened, asking if the new values should be adopted. This mechanism is turned off if the data have been coregistered to MRI (see online help chapter ''"MRI Coregistration''"), and an MRI Coregistration File (<span style="color:#ff9c00;">'''<nowiki>*.sfh</nowiki>'''</span>) has been associated with the data. | '''Function''': to redefine the center of the head for the sphere used in dipole models. If the cot file has the same base name as the data file, it is read automatically by BESA Research. If the coordinates deviate by more than 1 mm from the previously defined head center, a window is opened, asking if the new values should be adopted. This mechanism is turned off if the data have been coregistered to MRI (see online help chapter ''"MRI Coregistration''"), and an MRI Coregistration File (<span style="color:#ff9c00;">'''<nowiki>*.sfh</nowiki>'''</span>) has been associated with the data. | ||
| Line 186: | Line 189: | ||
'''Units:''' must be in meters! | '''Units:''' must be in meters! | ||
| − | + | ||
| + | Note: In special cases, a fifth value, the '''head radius''', may follow. This is used when reading simulated MEG data from the DipoleSimulator program. When this value is set, BESA Research uses the specified head radius and head center and does not fit a sphere to the head surface points. and does not create an ellipsoid transformation. | ||
Force BESA Research to use a completely spherical model without creating an ellipsoid: Write "'''DipoleSimulator"''' or "'''Phantom'''" on the second line of the file. Under these circumstances, 100% correspondence between DipoleSimulator and BESA Research is achieved. This is also required for dipole fitting on MEG phantom recordings. | Force BESA Research to use a completely spherical model without creating an ellipsoid: Write "'''DipoleSimulator"''' or "'''Phantom'''" on the second line of the file. Under these circumstances, 100% correspondence between DipoleSimulator and BESA Research is achieved. This is also required for dipole fitting on MEG phantom recordings. | ||
| − | The ''cot ''file has also been extended for reading CTF MEG files. Documentation for these extensions is found in the CTF help file. | + | The ''cot ''file has also been extended for reading CTF MEG files. Documentation for these extensions is found in the CTF help file. |
| + | == pos or pmg (MEG sensor coordinate) file == | ||
| − | + | '''Function:''' to define coordinates of MEG sensors. Our convention is to save magnetometer information in <span style="color:#ff9c00;"><tt>*.pmg</tt></span>, and gradiometer information in <span style="color:#ff9c00;"><tt>*.pos</tt></span>. In practice, BESA Research doesn’t mind which extension is used — the distinction between gradiometers and magnetometers is based on the number of values on each line in the file. | |
| − | + | ||
| − | '''Function:''' to define coordinates of MEG sensors. Our convention is to save magnetometer information in <span style="color:#ff9c00;"> | + | |
'''Format:''' one sensor per line. | '''Format:''' one sensor per line. | ||
| − | Magnetometers: label (optional), six coordinates per line (location, orientation) | + | |
| + | '''Magnetometers''' (<span style="color:#ff9c00;"><tt>*.pmg</tt></span>): label (optional), six coordinates per line (location, orientation) | ||
e.g. for BTi: | e.g. for BTi: | ||
| − | " Channel 'A1': -0.0019193 0.0304846 0.1081738 0.1188222 0.2394208 0.9636177 | + | <source lang="dos"> |
| + | Channel 'A1': -0.0019193 0.0304846 0.1081738 0.1188222 0.2394208 0.9636177 | ||
| + | </source> | ||
| − | Gradiometers: label (optional), nine coordinates per line (location of primary sensor, location of secondary sensor, orientation). The program decides whether gradiometers are planar or axial based on the distance between the primary and secondary sensor locations and the center of the head. | + | |
| + | '''Gradiometers''' (<span style="color:#ff9c00;"><tt>*.pos</tt></span>): label (optional), nine coordinates per line (location of primary sensor, location of secondary sensor, orientation). | ||
| + | |||
| + | The program decides whether gradiometers are planar or axial based on the distance between the primary and secondary sensor locations and the center of the head. | ||
e.g. for Neuromag: | e.g. for Neuromag: | ||
| − | " 0.108510 -0.000143 -0.044954 0.108510 0.000463 -0.028766 0.999999 0.001450 0.000000 | + | <source lang="dos"> |
| + | 0.108510 -0.000143 -0.044954 0.108510 0.000463 -0.028766 0.999999 0.001450 0.000000 | ||
| + | </source> | ||
Labels in these files are ignored. | Labels in these files are ignored. | ||
| − | See chapter | + | See chapter ''[[Electrodes_and_Surface_Locations#Data_reading_rules_for_MEG|Data reading rules for MEG]]''. |
| − | + | ||
| − | + | == sfn (surface point name) file == | |
'''Function:''' to match up digitized coordinates with channels that are defined as EEG electrodes and to define labels for additional digitized head surface points (e.g. MEG coils, etc.). | '''Function:''' to match up digitized coordinates with channels that are defined as EEG electrodes and to define labels for additional digitized head surface points (e.g. MEG coils, etc.). | ||
| Line 229: | Line 239: | ||
The case of labels is not important (e.g. 'Fp1' will match with 'fp1'). | The case of labels is not important (e.g. 'Fp1' will match with 'fp1'). | ||
| − | The ''sfn'' file need not exist if labels are defined in the ''sfp ''file. | + | The ''sfn'' file need not exist if labels are defined in the ''sfp'' file. |
| − | See chapter | + | See chapter ''[[Electrodes_and_Surface_Locations#Data_reading_rules_for_EEG|Data reading rules for EEG]]''. |
| − | + | == sfp (surface point coordinate) file == | |
| − | + | ||
'''Function''': to define coordinates of digitized points on the head surface. The order of points must match with the order in the ''sfn ''file, or if no ''sfn'' file is present, labels must be included in the ''sfp'' file. | '''Function''': to define coordinates of digitized points on the head surface. The order of points must match with the order in the ''sfn ''file, or if no ''sfn'' file is present, labels must be included in the ''sfp'' file. | ||
| Line 261: | Line 270: | ||
| − | + | == Generic File Format == | |
This reader, incorporated into the <span style="color:#ff9c00;">'''GenericBesa.dll'''</span> file, allows to read simple multiplexed or vectorized data formats, if you know the structure of the data format. | This reader, incorporated into the <span style="color:#ff9c00;">'''GenericBesa.dll'''</span> file, allows to read simple multiplexed or vectorized data formats, if you know the structure of the data format. | ||
| + | |||
'''What you have to do''' | '''What you have to do''' | ||
| + | |||
* With a text editor, write information about the data file you want to read into BESA Research into a text file, the ''Generic Header''. | * With a text editor, write information about the data file you want to read into BESA Research into a text file, the ''Generic Header''. | ||
* Save the edited text in the same subdirectory as the data file. | * Save the edited text in the same subdirectory as the data file. | ||
* '''Mechanism A:''' The generic header contains the data file name. With BESA Research, navigate to the file you just edited, and open it. The data should then be read into BESA Research. | * '''Mechanism A:''' The generic header contains the data file name. With BESA Research, navigate to the file you just edited, and open it. The data should then be read into BESA Research. | ||
* '''Mechanism B:''' Alternatively, navigate to the data file. The reader will check if there is a generic header in the same subdirectory, and use that to try to open the file. You have two options: | * '''Mechanism B:''' Alternatively, navigate to the data file. The reader will check if there is a generic header in the same subdirectory, and use that to try to open the file. You have two options: | ||
| − | + | ** '''Specific:''' if the file has the same basename as the data file, and the extension “<span style="color:#ff9c00;">'''.generic'''</span>”, this file will be used. | |
| − | * | + | ** '''Generalized:''' if a specific file is not found, the reader will look for the file “<span style="color:#ff9c00;">'''BESA.generic'''</span>” in the same subdirectory. |
| − | * | + | |
| − | + | ||
<span style="color:#ff0000;">'''Important note:'''</span><span style="color:#ff0000;"> We recommend using mechanism A, using a header with the extension "</span><span style="color:#ff9c00;"><span style="color:#ff0000;">'''.generic'''</span></span><span style="color:#ff0000;">". When opening the data file in BESA, select the generic header. Mechanism B sometimes fails when opening the data file in BESA Research, because one of the other readers in BESA may erroneously interpret the file as their "own" data format, sometimes leading to a crash.</span> | <span style="color:#ff0000;">'''Important note:'''</span><span style="color:#ff0000;"> We recommend using mechanism A, using a header with the extension "</span><span style="color:#ff9c00;"><span style="color:#ff0000;">'''.generic'''</span></span><span style="color:#ff0000;">". When opening the data file in BESA, select the generic header. Mechanism B sometimes fails when opening the data file in BESA Research, because one of the other readers in BESA may erroneously interpret the file as their "own" data format, sometimes leading to a crash.</span> | ||
| Line 284: | Line 293: | ||
Subsequent lines '''must''' contain the following parameters, in any order (note that the parameters are case insensitive): | Subsequent lines '''must''' contain the following parameters, in any order (note that the parameters are case insensitive): | ||
| − | + | {| class="wikitable" | |
| − | '''''nChannels | + | | width="20%" | '' '''nChannels''' = nnn '' || The number of channels |
| − | + | |- | |
| − | '''''sRate | + | | '' '''sRate''' = fff '' || The sampling rate (samples/sec) |
| − | + | |- | |
| − | '''''format | + | | '' '''format''' = type '' || One of ''short'', ''int, float, double, ASCII''. If the format is ''ASCII'', the parameter''' nSamples''' must be specified as well (see below) |
| − | + | |} | |
The following parameters are optional (values in square brackets denote optional parameters): | The following parameters are optional (values in square brackets denote optional parameters): | ||
| − | '''''nSamples | + | {| class="wikitable" |
| − | + | | width="15%" | '' '''nSamples'''= nnn '' || The number of time samples in the data. If this value is 0, or the line is omitted, then use the file size to estimate the number of samples. | |
| − | '''''file | + | |- |
| − | + | | '' '''file''' = || The data file name, without path information. If this is omitted, you can only read the data with mechanism B (see above). This line '''must''' be included if you want to read the data with mechanism A. | |
| − | '''''DataOffset | + | |- |
| − | + | | '' '''DataOffset''' = nnn'' || Offset of data in bytes for binary data, in lines for ASCII data (default = 0). | |
| − | '''''Factor | + | |- |
| − | + | | '' '''Factor''' = fff [range]'' || Data values are multiplied by this factor to obtain µV values (default = 1). Optional parameters can be appended to define a channel range, e.g. ''1-3''. Thus, this command can be used multiply, to define different scaling factors for different channels. If only one channel is specified, use on number only, e.g. ''5''. | |
| − | '''''SwapBytes | + | |- |
| − | + | | '' '''SwapBytes''' = ccc'' || One of ''off'' or ''on'' (default = off). If the data block originated from Unix or Mac, this will need to be ''on''. (binary data only) | |
| − | '''''Prestimulus | + | |- |
| − | + | | '' '''Prestimulus''' = fff'' || Prestimulus interval in milliseconds. | |
| − | '''''Label | + | |- |
| − | + | | '' '''Label''' = ccc'' || Segment label. | |
| − | '''''Trigger | + | |- |
| − | + | | '' '''Trigger''' = chan….'' || Channel number containing triggers. Without further parameters, the values are read directly as digital trigger values. Other parameters are described below, for the case where the trigger channel contains analog signals. | |
| − | '''''nBlocks | + | |- |
| − | + | | '' '''nBlocks''' = nnn'' || The data are epoched. This specifies the number of equal sized blocks in the data. In BESA Research, each block will be separated by a segment boundary. The number of samples in each epoch is computed from the total number of samples divided by ''nBlocks''. | |
| − | '''''nEpochs | + | |- |
| − | + | | '' '''nEpochs''' = nnn'' || Same as '''nBlocks'''. | |
| − | '''''EventFile | + | |- |
| − | + | | '' '''EventFile''' = name'' || Load events from an event file, using BESA's event file (<span style="color ||#ff9c00;">'''<nowiki>*.evt</nowiki>'''</span>) format. See below for a description of how to prepare this file. | |
| − | '''''Order | + | |- |
| − | + | | '' '''Order''' = type'' || One of ''multiplexed'', ''vectorized''. The default is multiplexed (i.e. channels fastest). Specify ''vectorized'' if your data are ordered so that all time samples for channel 1 are followed by all time samples from channel 2, etc. | |
| − | '''''Orientation | + | |- |
| − | + | | '' '''Orientation''' = type'' || Same as '''Order'''. | |
| − | '''''Arrangement | + | |- |
| + | | '' '''Arrangement''' = type'' || Same as '''Order'''. | ||
| + | |} | ||
| Line 332: | Line 343: | ||
'''Trigger''' = ''chan code fromLevel toLevel timerange'' ''deadtime'' | '''Trigger''' = ''chan code fromLevel toLevel timerange'' ''deadtime'' | ||
| − | '''''chan''' ''is the channel number on which to find the trigger | + | * '''''chan''' ''is the channel number on which to find the trigger |
| + | * '''''code''''' is the trigger number that the reader will assign (must be positive!) | ||
| + | * '''''fromLevel'' '''is the value in mV defining the lower range for trigger detection | ||
| + | * '''''toLevel'' '''is the value in mV defining the upper range for trigger detection. If this is “-“, then only ''fromLevel ''needs to be exceeded for the trigger to be detected. | ||
| + | * '''''timerange '''''is the range in milliseconds to define a trigger. The reader will search for the maximum deviation from baseline within the range to find the level that will define the trigger. | ||
| + | * '''''deadtime''' ''defines the time after detecting a trigger during which no further trigger with this code can be detected. This does not affect other codes. Also, if the voltage level stays at a level corresponding to a code, the trigger is only defined at the onset of this voltage level. | ||
| − | + | Multiple lines are required if different trigger codes and different trigger channels are required, one for each new code. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
'''Notes''' | '''Notes''' | ||
| Line 353: | Line 359: | ||
'''Data formats:''' | '''Data formats:''' | ||
| − | * Short 16-bit | + | * Short: 16-bit |
| − | * Int 32-bit | + | * Int: 32-bit |
| − | * Float 32-bit | + | * Float: 32-bit |
| − | * Double 64-bit | + | * Double: 64-bit |
| − | * ASCII Decimal numbers separated by spaces or tabs (engineering format also permitted) | + | * ASCII: Decimal numbers separated by spaces or tabs (engineering format also permitted) |
| − | + | ||
'''Prestimulus interval and label:''' If either of these are defined, BESA Research reads the data in to define an averaged data segment. The label is displayed, and a vertical dotted line marks timepoint zero. If no prestimulus interval is defined, a zero prestimulus interval is assumed. | '''Prestimulus interval and label:''' If either of these are defined, BESA Research reads the data in to define an averaged data segment. The label is displayed, and a vertical dotted line marks timepoint zero. If no prestimulus interval is defined, a zero prestimulus interval is assumed. | ||
| − | |||
'''Future changes''' | '''Future changes''' | ||
| Line 377: | Line 381: | ||
Each line contains four parameters: | Each line contains four parameters: | ||
| − | + | # latency (units specified by the header, can be µs, ms, s) | |
| − | + | # code (defines the type of event: trigger, comment, marker, pattern, average segment, data break segment) | |
| − | + | # parameter (depends on the event type, e.g. trigger code) | |
| − | + | # label (label assigned to the event) | |
| − | + | ||
| − | + | ||
| − | + | ||
| Line 390: | Line 391: | ||
The header line contains four values. The first specifies the time units, e.g. '''Tmu '''specifies microseconds. | The header line contains four values. The first specifies the time units, e.g. '''Tmu '''specifies microseconds. | ||
| − | + | <source lang="dos"> | |
| − | + | Tmu Code TriNo Comnt | |
| + | </source> | ||
'''Tms''' specifies milliseconds. '''Tsec''' specifies seconds. | '''Tms''' specifies milliseconds. '''Tsec''' specifies seconds. | ||
| Line 418: | Line 420: | ||
41 = segment onset -- '''TriNo '''is a time string that specifies date and time, in the format ''YYYY-MM-DDTHH:MM:SS'', e.g. ''2010-04-26T15:30:20.31'' (note: seconds are a decimal number). | 41 = segment onset -- '''TriNo '''is a time string that specifies date and time, in the format ''YYYY-MM-DDTHH:MM:SS'', e.g. ''2010-04-26T15:30:20.31'' (note: seconds are a decimal number). | ||
| − | 42 = average segment onset -- '''TriNo '''is a number specifying the prestimulus baseline of the subsequent average in microseconds. '''TriNo '''(parameter) is 0 for markers, comments, artifacts, and epochs. | + | 42 = average segment onset -- '''TriNo '''is a number specifying the prestimulus baseline of the subsequent average in microseconds. |
| + | |||
| + | '''TriNo '''(parameter) is 0 for markers, comments, artifacts, and epochs. | ||
| Line 430: | Line 434: | ||
A simple way to generate example files is to export events from BESA (''ERP/Save Events As...''). | A simple way to generate example files is to export events from BESA (''ERP/Save Events As...''). | ||
| − | Tmu | + | <source lang="dos"> |
| − | + | Tmu Code TriNo Comnt | |
| − | 0 | + | 0 42 100000 Ave: 25 avs |
| − | + | 10000000 2 0 Comment at 10s | |
| − | 10000000 | + | 20000000 41 26-04-2010T15:30:20.000 TestSeg2 |
| − | + | 21000000 3 0 | |
| − | 20000000 | + | 22000000 1 99 Trigger - 99 |
| − | + | </source> | |
| − | 21000000 | + | |
| − | + | ||
| − | 22000000 1 | + | |
| − | + | ||
This specifies an average segment starting at the beginning of the file, with a prestimulus interval of 100 ms, a comment at 10 s, a new segment specifying date and time at 20 s, a marker at 21 s, and a trigger with code 99 at 22 s. | This specifies an average segment starting at the beginning of the file, with a prestimulus interval of 100 ms, a comment at 10 s, a new segment specifying date and time at 20 s, a marker at 21 s, and a trigger with code 99 at 22 s. | ||
| Line 450: | Line 450: | ||
The following reads ASCII multiplexed data that were previously exported from BESA Research: | The following reads ASCII multiplexed data that were previously exported from BESA Research: | ||
| − | + | <source lang="dos"> | |
| − | + | BESA Generic Data | |
| − | + | nchannels = 64 | |
| − | + | srate = 100 | |
| − | + | nsamples = 10000 | |
| − | + | dataoffset = 2 | |
| − | + | format = ASCII | |
| − | + | file = name.mul | |
| − | + | factor = 1 | |
| − | + | </source> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
With the sampling rate of 100 Hz and 10000 samples, this represents 100 s of 64-channel data. The first two lines of the data are skipped. | With the sampling rate of 100 Hz and 10000 samples, this represents 100 s of 64-channel data. The first two lines of the data are skipped. | ||
Latest revision as of 12:40, 5 May 2021
| Module information | |
| Modules | BESA Research Basic or higher |
| Version | BESA Research 6.1 or higher |
Contents
- 1 Binary format (*.foc, *.fsg)
- 2 ASCII vectorized format (*.avr)
- 3 ASCII Multiplexed format (*.mul)
- 4 Channel definition file conventions and formats
- 5 cot (Head center) file
- 6 pos or pmg (MEG sensor coordinate) file
- 7 sfn (surface point name) file
- 8 sfp (surface point coordinate) file
- 9 Generic File Format
Binary format (*.foc, *.fsg)
Select Binary High Resolution or Binary Compressed Format to output segments in binary BESA format. If the file already exists, the segment will be appended. Thus, it is possible to create a file combining several segments of interest in a compact form. BESA Research will only allow you to append segments if the number of channels and the sampling interval in source and target files are the same.
In Binary Format all channels (scalp, intracranial, polygraphic, MEG) and file events in the selected time range are exported. Note: The channels are filtered according to the current filter settings.
Select Binary High Resolution to retain the resolution of the processed data. This is the preferred binary format for small amplitude signals such as averages. Select Binary Compressed Format to store raw data using the original resolution or to obtain the space savings of compression (see File/Export and Append Data/Convert.. above). This is the preferred binary format for raw data.
ASCII vectorized format (*.avr)
Select ASCII vectorized Format to output segments in BESA ASCII format, one channel (all time points) per line.
The Format is as follows:
The first of two header lines contains the following data descriptors (6 descriptors, the values shown are only examples):
| Npts= 200 | number of sampled points in each channel |
| TSB= -500 | time sweep begin [ms]. Time of first data point relative to zero of epoch |
| DI= 5 | digitization or sampling interval [ms] |
| SB= 2 | scaling bins/microvolt in file = number corresponding to 1 microvolt |
| SC= 50 | scaling calibration, used for setting magnitude of display in BESA |
| Nchan= 27 | number of channels |
| SegmentName= 60dB | an optional label describing the data |
The second line of the header contains a label for each channel, e.g.
O1 Oz P3 T5 T3 C3 F7 F3 Fp1 Fz Cz Pz Fp2 F4 F8 C4 T4 T6 P4 Fpz O2 M2 M1 F10 F9 T10 T9
Each of the subsequent Nchan lines of the file contains values for all Npts time points in floating point or scientific format. For more details about scalp electrodes, see chapter Working With Electrodes and Surface Locations/Electrodes/Electrode Conventions.
A second (older) version of the format (written by BESA versions 1, 2 and 3) omits the Nchan=xx information in the first line, and there is no second header line. Labels must be defined elsewhere. See Channel definition file conventions and formats and Reading MEG files in ASCII format. In the older versions, only scalp channels were exported, and the data were average referenced.
ASCII Multiplexed format (*.mul)
Select ASCII Multiplexed Format to output segments one time point (all channels) per line.
The ASCII Multiplexed Format is as follows:
The first of two header lines contains similar information to that of the ASCII vectorized file:
TimePoints= 200 Channels= 27 BeginSweep[ms]= -500.00 SamplingInterval[ms]= 5.000 Bins/uV= 1.000 SegmentName=Condition1
Note that the item SegmentName is missing if no segment comment is specified when writing a segment to file.
If an epoch of a continuous EEG is exported in ASCII multiplexed format, the first header line contains the additional item Time, which indicates the daytime of the first sample in the exported segment:
TimePoints= 200 Channels= 27 BeginSweep[ms]= 0.00 SamplingInterval[ms]= 5.000 Bins/uV= 1.000 Time=22:02:53 SegmentName=Segment1
The second line of the header contains labels for each channel, which may be either the original channel names, or the names of the channels of the current montage, e.g.
O1 Oz P3 T5 T3 C3 F7 F3 Fp1 Fz Cz Pz Fp2 F4 F8 C4 T4 T6 P4 Fpz O2 M2 M1 F10 F9 T10 T9
Each subsequent line contains values for all 'Channels' at one time point, in floating point or scientific format. Values are given for the current or the original montage, selected as described above.
Labels for source montages have the following form: TAr-L.
- The first two letters indicate the head region:
- TA: temporal-anterior
- TP: temporal-posterior
- TB: temporal-basal
- FL: fronto-lateral
- FP: fronto-polar
- PC: posterior-central
- OL: occipito-lateral
- OC: occipito-central
- CL: centro-lateral
- CC: central
- The small letter indicates in part the orientation: r=radial, t=tangential, and in part the relative location of the basal temporal source: l=lateral, m=mesial.
- The final letter after the hyphen indicates L=left, M=middle, R=right.
Channel definition file conventions and formats
BESA Research can read 3 types of file to define channels. These are identified by different extensions:
- *.ela: channel definition files containing labels and, optionally, channel types (ASCII, 1 label / line)
- *.elp: channel definition files containing coordinates and, optionally, channel types and labels (ASCII). This is the format of the file written by the Channel Configuration Dialog.
- *.elb: channel definition files stored by older versions of BESA Research (binary format). This format can still be read, but is no longer written by BESA Research.
BESA Research stores and retrieves the channel configuration after editing in binary files. If you open a data file, BESA Research will search for the related channel information in the following sequence:
- For all data files with the extension .eeg, .cnt and .foc, check in the additional database file in the data directory with the same basename as the data file and the extension .fst, whether a channel file has been associated previously
- Check in the db subdirectory whether a channel file has been associated previously
- Check if labels are defined in the header of the data file
- Search for a corresponding binary channel definition file with the same basename as the data file in the data folder (xxxx.elb)
- Search for a corresponding channel definition file with the same basename as the data file containing labels in the data folder (xxxx.ela)
- Search for a corresponding channel definition file with the same basename as the data file containing labels and/or coordinates in the data folder (xxxx.elp)
- Search for a file named default.elb, default.ela or default.elp (in this order)) in the data folder
- Search for a file named default.elb, default.ela or default.elp one directory above the data folder (e.g. ..\default.elb)
- Check if the new data file is of the same type and has the same number of channels as the preceding data file in the list. If this is the case, the electrode configuration of the previous file will be assumed. This will avoid having to load or edit the electrode configuration more than once, if you load several data segments of the same subject from separate files.
- If no channel definition file is found, or digitization points (in files *.sfp, *.cot, *.pos, *.pmg) are found in files with the same basename as the data file, the "Channel and digitized surface point information" dialog box is opened, allowing you to specify file names.
Channel Label Files (*.ela)
Files containing a list of channel labels are an alternative to editing electrode configurations. They can be edited using a standard text editor. Electrode label files (.ela) require a sequence of lines corresponding to the sequence of channels in the data. Each line contains one label and an optional identifier. The format is ' [Identifier] {Label} ', ('Identifier' can be omitted if the electrode label defines the type of signal)
Identifiers can be one of:
- EEG - scalp electrode
- SCP - scalp electrode
- POL - polygraphic channel
- PGR - polygraphic channel
- ICR - intracranial electrode
- MEG - MEG sensor
- REF - reference electrode (this can only occur once, and must be the last item in the file)
For example:
Fz (scalp electrode, coordinates assigned by default.ecd)
Cz (scalp electrode, coordinates assigned by default.ecd)
......
VEOG (vertical EOG, Polygraphic type is assigned by default)
Exx (xx=01, 02.. electrode number, Polygraphic type is assigned by default)
......
EEG xx (scalp electrode, coordinates must be assigned either by default.ecd or by a surface point (+.sfp) file. An alternative to the "EEG" prefix is "SCP".
......
POL XX (identifier sets Polygraphic type -- an alternative to the "POL" prefix is "PGR")
......
ICR A01 (identifier sets Intracranial type to electrode A01 - do not use A1!)
ICR A02 (identifier sets Intracranial type to electrode A02 - do not use A2!)
ICR A03 (identifier sets Intracranial type to electrode A03)
......
MEG M01 (identifier sets MEG type to electrode M01 - do not use M1!)
MEG M02 (identifier sets MEG type to electrode M02 - do not use M2!)
......
REF Cz (the label is assigned to the electrode reference, no channel is associated with this entry. This must be the last line of the file!)
Channel spherical coordinate files (*.elp)
These files follow the same rules as the channel label files, with the addition of spherical coordinates (theta, phi) that follow the labels of EEG and MEG channels. Labels can also be omitted. In this case, BESA Research will assign labels according to the nearest coordinate defined in the default.ecd file. To indicate that the coordinates have been assigned, the label will have a tick, e.g. Fz' instead of Fz.
Channels of other types (polygraphic, intracranial) are defined exactly as in the channel label (*.ela) file.
cot (Head center) file
Function: to redefine the center of the head for the sphere used in dipole models. If the cot file has the same base name as the data file, it is read automatically by BESA Research. If the coordinates deviate by more than 1 mm from the previously defined head center, a window is opened, asking if the new values should be adopted. This mechanism is turned off if the data have been coregistered to MRI (see online help chapter "MRI Coregistration"), and an MRI Coregistration File (*.sfh) has been associated with the data.
BESA Research uses any head surface points (e.g. electrode locations), excluding those on the lower part of the face, to compute the sphere center automatically. The cot file is used if you want to override the automatic calculation. A mechanism is provided which allows to pass a location from the MRI (viewed by BrainVoyager) to the Source Module and save the resulting location as a cot file.
Format: one set of coordinates (x y z). These are followed by either "DC" or "HC", which specify whether these coordinates are in Device or Head Coordinates.
Units: must be in meters!
Note: In special cases, a fifth value, the head radius, may follow. This is used when reading simulated MEG data from the DipoleSimulator program. When this value is set, BESA Research uses the specified head radius and head center and does not fit a sphere to the head surface points. and does not create an ellipsoid transformation.
Force BESA Research to use a completely spherical model without creating an ellipsoid: Write "DipoleSimulator" or "Phantom" on the second line of the file. Under these circumstances, 100% correspondence between DipoleSimulator and BESA Research is achieved. This is also required for dipole fitting on MEG phantom recordings.
The cot file has also been extended for reading CTF MEG files. Documentation for these extensions is found in the CTF help file.
pos or pmg (MEG sensor coordinate) file
Function: to define coordinates of MEG sensors. Our convention is to save magnetometer information in *.pmg, and gradiometer information in *.pos. In practice, BESA Research doesn’t mind which extension is used — the distinction between gradiometers and magnetometers is based on the number of values on each line in the file.
Format: one sensor per line.
Magnetometers (*.pmg): label (optional), six coordinates per line (location, orientation)
e.g. for BTi:
Channel 'A1': -0.0019193 0.0304846 0.1081738 0.1188222 0.2394208 0.9636177
Gradiometers (*.pos): label (optional), nine coordinates per line (location of primary sensor, location of secondary sensor, orientation).
The program decides whether gradiometers are planar or axial based on the distance between the primary and secondary sensor locations and the center of the head.
e.g. for Neuromag:
0.108510 -0.000143 -0.044954 0.108510 0.000463 -0.028766 0.999999 0.001450 0.000000
Labels in these files are ignored.
See chapter Data reading rules for MEG.
sfn (surface point name) file
Function: to match up digitized coordinates with channels that are defined as EEG electrodes and to define labels for additional digitized head surface points (e.g. MEG coils, etc.).
Format: one label per line.
Contains labels of surface points in the order of digitization. If fiducials are defined, these should be on the first three lines, with the labels 'FidT9', 'FidT10', 'FidNz' or 'FidLPA', 'FidRPA', 'FidNAS'.
If electrodes are defined in the data file, the labels of each electrode as defined in the data file (or in its associated ela, elp, or elb file) must be present!
The case of labels is not important (e.g. 'Fp1' will match with 'fp1').
The sfn file need not exist if labels are defined in the sfp file.
See chapter Data reading rules for EEG.
sfp (surface point coordinate) file
Function: to define coordinates of digitized points on the head surface. The order of points must match with the order in the sfn file, or if no sfn file is present, labels must be included in the sfp file.
Note: If the digitized points include electrodes, the channel labels must correspond to the labels of the digitized points. The sequence of labels in channels and surface point coordinate file need not be the same – the allocation is performed by label matching. Channel labels may be defined in the data file, or they may be assigned using channel definition files (*.ela, *.elp, *.elb).
Format: one set of coordinates (x, y, z) per line. Coordinate units must be either meter, centimeter or millimeter (BESA Research will perform a plausibility check automatically to determine which units are used). If a label is present this can precede or come after the three coordinate values.
If fiducials are defined, these should be on the first three lines. BESA Research will simulate fiducials if none are defined, but it is preferable to record these locations along with the other head surface points.
If there are MEG sensors, the same coordinate systems must be used in the sfp file and the pos/pmg file!
If labels are defined in the sfp file rather than in an sfn file, labeling rules apply as for the sfn file.
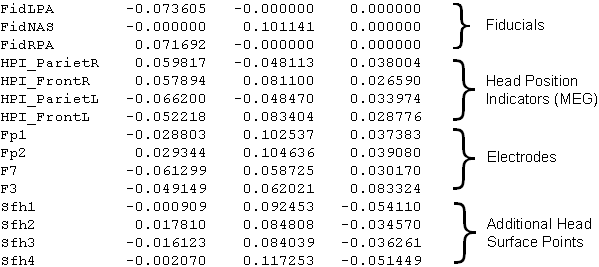
Example for the sfp file format:
BESA Research will check the coordinates for plausibility. If coordinates are more than 30° away from the expected location on the sphere there will be an error message. Such errors are usually due either to incorrect labeling or to a digitization error.
See chapter “3D Coordinates for Precise Analysis / Data reading rules for EEG”.
Generic File Format
This reader, incorporated into the GenericBesa.dll file, allows to read simple multiplexed or vectorized data formats, if you know the structure of the data format.
What you have to do
- With a text editor, write information about the data file you want to read into BESA Research into a text file, the Generic Header.
- Save the edited text in the same subdirectory as the data file.
- Mechanism A: The generic header contains the data file name. With BESA Research, navigate to the file you just edited, and open it. The data should then be read into BESA Research.
- Mechanism B: Alternatively, navigate to the data file. The reader will check if there is a generic header in the same subdirectory, and use that to try to open the file. You have two options:
- Specific: if the file has the same basename as the data file, and the extension “.generic”, this file will be used.
- Generalized: if a specific file is not found, the reader will look for the file “BESA.generic” in the same subdirectory.
Important note: We recommend using mechanism A, using a header with the extension ".generic". When opening the data file in BESA, select the generic header. Mechanism B sometimes fails when opening the data file in BESA Research, because one of the other readers in BESA may erroneously interpret the file as their "own" data format, sometimes leading to a crash.
Format of the Generic Header
The first line must consist of the text: “BESA Generic Data” (without the inverted commas).
Subsequent lines must contain the following parameters, in any order (note that the parameters are case insensitive):
| nChannels = nnn | The number of channels |
| sRate = fff | The sampling rate (samples/sec) |
| format = type | One of short, int, float, double, ASCII. If the format is ASCII, the parameter nSamples must be specified as well (see below) |
The following parameters are optional (values in square brackets denote optional parameters):
| nSamples= nnn | The number of time samples in the data. If this value is 0, or the line is omitted, then use the file size to estimate the number of samples. |
| file = | The data file name, without path information. If this is omitted, you can only read the data with mechanism B (see above). This line must be included if you want to read the data with mechanism A. |
| DataOffset = nnn | Offset of data in bytes for binary data, in lines for ASCII data (default = 0). |
| Factor = fff [range] | Data values are multiplied by this factor to obtain µV values (default = 1). Optional parameters can be appended to define a channel range, e.g. 1-3. Thus, this command can be used multiply, to define different scaling factors for different channels. If only one channel is specified, use on number only, e.g. 5. |
| SwapBytes = ccc | One of off or on (default = off). If the data block originated from Unix or Mac, this will need to be on. (binary data only) |
| Prestimulus = fff | Prestimulus interval in milliseconds. |
| Label = ccc | Segment label. |
| Trigger = chan…. | Channel number containing triggers. Without further parameters, the values are read directly as digital trigger values. Other parameters are described below, for the case where the trigger channel contains analog signals. |
| nBlocks = nnn | The data are epoched. This specifies the number of equal sized blocks in the data. In BESA Research, each block will be separated by a segment boundary. The number of samples in each epoch is computed from the total number of samples divided by nBlocks. |
| nEpochs = nnn | Same as nBlocks. |
| EventFile = name | Load events from an event file, using BESA's event file (*.evt) format. See below for a description of how to prepare this file. |
| Order = type | One of multiplexed, vectorized. The default is multiplexed (i.e. channels fastest). Specify vectorized if your data are ordered so that all time samples for channel 1 are followed by all time samples from channel 2, etc. |
| Orientation = type | Same as Order. |
| Arrangement = type | Same as Order. |
Trigger events
This section describes how the reader can be used to encode trigger events when the trigger channel contains analog signals. In this case, a Trigger command is required for each target code in BESA Research.
Syntax:
Trigger = chan code fromLevel toLevel timerange deadtime
- chan is the channel number on which to find the trigger
- code is the trigger number that the reader will assign (must be positive!)
- fromLevel is the value in mV defining the lower range for trigger detection
- toLevel is the value in mV defining the upper range for trigger detection. If this is “-“, then only fromLevel needs to be exceeded for the trigger to be detected.
- timerange is the range in milliseconds to define a trigger. The reader will search for the maximum deviation from baseline within the range to find the level that will define the trigger.
- deadtime defines the time after detecting a trigger during which no further trigger with this code can be detected. This does not affect other codes. Also, if the voltage level stays at a level corresponding to a code, the trigger is only defined at the onset of this voltage level.
Multiple lines are required if different trigger codes and different trigger channels are required, one for each new code.
Notes
Channel labels: The data channels are labeled E1, E2, E3,…, and they are initially classified by BESA Research as polygraphic. As with other BESA Research data files, use a *.ela file (or *.elp, optionally combined with *.sfp) to redefine labels and channel types.
Data formats:
- Short: 16-bit
- Int: 32-bit
- Float: 32-bit
- Double: 64-bit
- ASCII: Decimal numbers separated by spaces or tabs (engineering format also permitted)
Prestimulus interval and label: If either of these are defined, BESA Research reads the data in to define an averaged data segment. The label is displayed, and a vertical dotted line marks timepoint zero. If no prestimulus interval is defined, a zero prestimulus interval is assumed.
Future changes
Possible developments:
- Read channel labels
If any of these changes are particularly important to you, please contact support@besa.de and let us know.
Event File
The event file is a text (ASCII) file containing a header line and subsequent lines, with one event description per line.
Each line contains four parameters:
- latency (units specified by the header, can be µs, ms, s)
- code (defines the type of event: trigger, comment, marker, pattern, average segment, data break segment)
- parameter (depends on the event type, e.g. trigger code)
- label (label assigned to the event)
Header Line:
The header line contains four values. The first specifies the time units, e.g. Tmu specifies microseconds.
Tmu Code TriNo Comnt
Tms specifies milliseconds. Tsec specifies seconds.
Event Code and Parameter 3 (TriNo):
Code specifies the event type:
1 = trigger -- TriNo specifies the trigger number
2 = comment
3 = marker
11-15 = patterns 1-5
21 = artifact on
22 = artifact off
31 = epoch on
32 = epoch off
41 = segment onset -- TriNo is a time string that specifies date and time, in the format YYYY-MM-DDTHH:MM:SS, e.g. 2010-04-26T15:30:20.31 (note: seconds are a decimal number).
42 = average segment onset -- TriNo is a number specifying the prestimulus baseline of the subsequent average in microseconds.
TriNo (parameter) is 0 for markers, comments, artifacts, and epochs.
Comment
The event label. This is not used for markers, artifacts, and epochs.
Example of event file:
A simple way to generate example files is to export events from BESA (ERP/Save Events As...).
Tmu Code TriNo Comnt 0 42 100000 Ave: 25 avs 10000000 2 0 Comment at 10s 20000000 41 26-04-2010T15:30:20.000 TestSeg2 21000000 3 0 22000000 1 99 Trigger - 99
This specifies an average segment starting at the beginning of the file, with a prestimulus interval of 100 ms, a comment at 10 s, a new segment specifying date and time at 20 s, a marker at 21 s, and a trigger with code 99 at 22 s.
Examples
The following reads ASCII multiplexed data that were previously exported from BESA Research:
BESA Generic Data nchannels = 64 srate = 100 nsamples = 10000 dataoffset = 2 format = ASCII file = name.mul factor = 1
With the sampling rate of 100 Hz and 10000 samples, this represents 100 s of 64-channel data. The first two lines of the data are skipped.
| Review | |
|---|---|
| Source Analysis | |
| Integration with MRI and fMRI | |
| Source Coherence | |
| Export | |
| MATLAB Interface | |
| Special Topics |
|